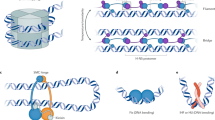
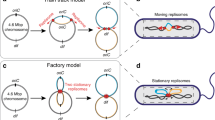
Abstract
We present a new physical biology approach to understanding the relationship between the organization and segregation of bacterial chromosomes. We posit that replicated Escherichia coli daughter strands will spontaneously demix as a result of entropic forces, despite their strong confinement within the cell; in other words, we propose that entropy can act as a primordial physical force which drives chromosome segregation under the right physical conditions. Furthermore, proteins implicated in the regulation of chromosome structure and segregation may in fact function primarily in supporting such an entropy-driven segregation mechanism by regulating the physical state of chromosomes. We conclude that bacterial chromosome segregation is best understood in terms of spontaneous demixing of daughter strands. Our concept may also have important implications for chromosome segregation in eukaryotes, in which spindle-dependent chromosome movement follows an extended period of sister chromatid demixing and compaction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Kornberg, A. & Baker, T. A. DNA Replication 2nd edn (Freeman & Company, New York, 1992).
Murray, A. & Hunt, T. (eds) The Cell Cycle: an Introduction. (Oxford Univ. Press, New York, 1993).
Wang, J. C. DNA topoisomerases. Annu. Rev. Biochem. 65, 635–692 (1996).
Sullivan, N. L., Marquis, K. A. & Rudner, D. Z. Recruitment of SMC to the origin by ParB-parS organizes the origin and promotes efficient chromosome segregation. Cell 137, 697–707 (2009).
Gruber, S. & Errington, J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137, 685–696 (2009).
Salje, J., Zuber, B. & Löwe, J. Electron cryomicroscopy of E. coli reveals ParM filament bundles involved in plasmid segregation. Science 323, 509–512 (2009).
deGennes, P.-G. Scaling Concepts in Polymer Physics (Cornell Univ. Press, Ithaca, New York, 1979).
Bloom, K. & Joglekar, A. Towards building a chromosome segregation machine. Nature 463, 446–456 (2010).
Stavans, J. & Oppenheim, A. DNA-protein interactions and bacterial chromosome architecture. Phys. Biol. 3, R1–R10 (2006).
Woldringh, C. L. & Odijk, T. in Organization of the Prokaryotic Genome (ed. Charlebois, R. L.) 171–187 (American Society for Microbiology, Washington DC, 1999).
Romantsov, T., Fishov, I. & Krichevsky, O. Internal structure and dynamics of isolated Escherichia coli nucleoids assessed by fluorescence correlation spectroscopy. Biophys. J. 92, 2875–2884 (2007).
Derman, A. I., Lim-Fong, G. & Pogliano, J. Intracellular mobility of plasmid DNA is limited by the ParA family of partitioning systems. Mol. Microbiol. 67, 935–946 (2008).
Jun, S., Arnold, A. & Ha, B.-Y. Confined space and effective interactions of multiple self-avoiding chains. Phys. Rev. Lett. 98, 128303 (2007).
Sawitzke, J. A. & Austin, S. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc. Natl Acad. Sci. USA 97, 1671–1676 (2000).
Jun, S. & Mulder, B. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc. Natl Acad. Sci. USA 103, 12388–12393 (2006).
Trun, N. J. & Marko, J. F. Architecture of a bacterial chromosome. ASM News 64, 276–283 (1998).
Vilgis, T. A. Polymer theory: path integrals and scaling. Phys. Rep. 336, 167–254 (2000).
Huang, J. C. & Wingreen, N. Min-protein oscillations in round bacteria. Phys. Biol. 1, 229–235 (2004).
Corbin, B. D., Yu, X.-C. & Margolin, W. Exploring intracellular space: function of the Min system in round-shaped Escherichia coli. EMBO J. 21, 1998–2008 (2002).
Berlatzky, I. A., Rouvinski, A. & Ben-Yehuda, S. Spatial organization of a replicating bacterial chromosome. Proc. Natl Acad. Sci. USA 105, 14136–14140 (2008).
Nordstrom, K. & Austin, S. J. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 23, 37–69 (1989).
Gordon, G. S. et al. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell 90, 1113–1121 (1997).
Danilova, O., Reyes-Lamothe, R., Pinskaya, M., Sherratt, D. & Possoz, C. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol. Microbiol. 65, 1485–1492 (2007).
Viollier, P. H., Thanbichle, M., McGrath, P. T., West, L. & Meewan, M. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl Acad. Sci. USA 101, 9257–9262 (2004).
Fogel, M. A. & Waldor, M. K. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 20, 3269–3282 (2006).
Fiebig, A., Keren, K. & Theriot, J. A. Fine-scale time-lapse analysis of the biphasic, dynamic behaviour of the two Vibrio cholerae chromosomes. Mol. Microbiol. 60, 1164–1178 (2006).
Toro, E., Hong, S.-H., McAdams, H. H. & Shapiro, L. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc. Natl Acad. Sci. USA 105, 15435–15440 (2008).
Nasmyth, K. Segregating sister genomes: the molecular biology of chromosome separation. Science 297, 559–565 (2002).
Barre, F.-X. FtsK and SpoIIIE: the tale of the conserved tails. Mol. Microbiol. 66, 1051–1055 (2007).
Lemon, K. P. & Grossman, A. D. The extrusion capture model for chromosome partitioning in bacteria. Genes Dev. 15, 2031–2041 (2001).
Migocki, M. D., Lewis, P. J., Wake, R. G. & Harry, E. J. The midcell replication factory in Bacillus subtilis is highly mobile: implications for coordinating chromosome replication with other cell cycle events. Mol. Microbiol. 54, 452–463 (2004).
Bates, D. The bacterial replisome: back on track? Mol. Microbiol. 69, 1341–1348 (2008).
Reyes-Lamothe, R., Possoz, C., Danilova, O. & Sherratt, D. J. Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133, 90–102 (2008).
Dworkin, J. & Losick, R. Does RNA polymerase help drive chromosome segregation in bacteria? Proc. Natl Acad. Sci. USA 99, 14089–14094 (2002).
Woldringh, C. L. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol. Microbiol. 45, 17–29 (2002).
Fan, J., Tuncay, K. & Ortoleva, P. J. Chromosome segregation in Escherichia coli division: a free energy-driven string model. Comput. Biol. Chem. 31, 257–264 (2007).
Holmes, V. F. & Cozzarelli, N. R. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc. Natl Acad. Sci. USA 97, 1322–1324 (2000).
Dasgupta, S., Maisnier-Patin, S. & Nordstrom, K. New genes with old modus operandi: the connection between supercoiling and partitioning of DNA in Escherichia coli. EMBO Rep. 1, 323–327 (2000).
Weart, R. B. et al. A metabolic sensor governing cell size in bacteria. Cell 130, 335–347 (2007).
Madabhushi, R. & Marians, K. J. Active homolog MreB affects chromosome segregation by regulating topoisomerase IV in Escherichia coli. Mol. Cell 33, 171–180 (2009).
Wang, X., Liu, X., Possoz, C. & Sherratt, D. J. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 20, 1727–1731 (2006).
Nielsen, H. J., Ottensen, J. R., Youngren, B., Austin, S. J. & Hansen, F. G. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol. Microbiol. 62, 331–338 (2006).
Nielsen, H. J., Li, Y., Youngren, B., Hansen, F. G. & Austin, S. J. Progressive segregation of the Escherichia coli chromosome. Mol. Microbiol. 61, 383–393 (2006).
Wiggins, P. A., Cheveralls, K., Martin, J. S., Lintner, R. & Kondev, J. Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc. Natl Acad. Sci. USA 107, 4991–4995 (2010)
White, M. A., Eykelenboom, J. K., Lopez-Vernaza, M. A., Wilson, E. & Leach, D. R. F. Non-random segregation of sister chromosomes in Escherichia coli. Nature 455, 1248–1250 (2008).
Reyes-Lamothe, R., Wang, X. & Sherratt, D. Escherichia coli and its chromosome. Trends Microbiol. 16, 238–245 (2008).
Kleckner, N. et al. A mechanical basis for chromosome function. Proc. Natl Acad. Sci. USA 101, 12592–12597 (2004).
Bates, D. & Kleckner, N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121, 899–911 (2005).
Woldringh, C. L. & van Driel, R. in Organization of the Prokaryotic Genome Ch. 5 (ed. Charlebois, R. L.) 77–90 (American Society for Microbiology, Washington DC, 1999).
Schrödinger, E. What is life? (Cambridge Univ. Press, Cambridge, UK, 1944).
Flory, P. Principles of Polymer Chemistry (Cornell Univ. Press, Ithaca, New York, 1953).
Grosberg, A. Y., Khalatur, P. G. & Khokhloo, A. R. Polymeric coils with excluded volume in dilute solution: the invalidity of the model of impenetrable spheres and the influence of excluded volume on the rates of diffusion-controlled intermacromolecular reactions. Makromol. Chem. Rapid Commun. 3, 709–713 (1982).
Pincus, P. Excluded volume effects and stretched polymer chains. Macromolecules 9, 386–388 (1976).
Liu, Z., Zechiedrich, E. L. & Chan, H. S. Inferring global topology from local juxtaposition geometry: interlinking polymer rings and ramifications for topoisomerase action. Biophys. J. 90, 2344–2355 (2006).
Jacob, F., Brenner, S. & Cuzin, F. On the regulation of DNA replication in bacteria. Cold Spring Harb. Symp. Quant. Biol. 28, 329–348 (1963).
Kruse, T. & Gerdes, K. Bacterial DNA segregationby the actin-like MreB protein. Trends. Cell Biol. 15, 343–345 (2005).
Hu, B., Yang, G., Zhao, W., Zhang, Y. & Zhao, J. MreB is important for cell shape but not for chromosome segregation of the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 63, 1640–1652 (2007).
Karczmarek, A. et al. DNA and origin region segregation are not affected by the transition from rod to sphere after inhibition of Escherichia coli MreB by A22. Mol. Microbiol. 65, 51–63 (2007).
Yamaichi, Y. & Niki, H. migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J. 23, 221–233 (2004).
Niki, H., Jaffe, A., Imamura, R., Ogura, T. & Hiraga, S. The new gene mukB codes for a 177 kD protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 10, 183–193 (1991).
Acknowledgements
We are deeply grateful to T. Mitchison for his penetrating insight and numerous invaluable suggestions. We thank N. Kleckner and C. Woldringh for critical reading of the manuscript. We also thank J.-Y. Bouet, A. Brouniquel, A. Danchin, E. Garner, B.-Y. Ha, R. Losick, K. Maeshima, B. Mulder, A. Murray, P. Wiggins and many other colleagues for helpful discussions over the years. This work was supported by Harvard University, USA, and the US National Institutes of Health (grant P50 GM068763 to S.J.).
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (movie)
(MP4 2015 kb)
Supplementary information S2 (movie)
(MP4 2394 kb)
Supplementary information S3 (box)
(PDF 909 kb)
Supplementary information S4 (box)
(PDF 3950 kb)
Related links
Related links
DATABASES
Entrez Genome Project
FURTHER INFORMATION
Glossary
- Contour length
-
The length of the polymer at maximum extension.
- Ideal gas
-
A theoretical gas consisting of randomly-moving, non-interacting point particles.
Rights and permissions
About this article
Cite this article
Jun, S., Wright, A. Entropy as the driver of chromosome segregation. Nat Rev Microbiol 8, 600–607 (2010). https://doi.org/10.1038/nrmicro2391
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2391
This article is cited by
-
Modeling the compaction of bacterial chromosomes by biomolecular crowding and the cross-linking protein H-NS
Scientific Reports (2024)
-
Breaking the bottleneck of synthetic cells
Nature Nanotechnology (2024)
-
Mid-cell migration of the chromosomal terminus is coupled to origin segregation in Escherichia coli
Nature Communications (2023)
-
Confinement anisotropy drives polar organization of two DNA molecules interacting in a nanoscale cavity
Nature Communications (2022)
-
Towards a synthetic cell cycle
Nature Communications (2021)