Key Points
-
The stringent response is a regulatory mechanism that is controlled by members of the RelA–SpoT homologue (RSH) protein family in response to stress in bacteria. It is mediated by two related alarmone nucleotides, guanosine tetraphosphate and guanosine pentaphosphate, which are collectively referred to as (p)ppGpp.
-
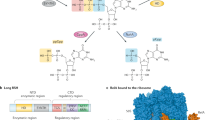
The RSH enzymes can be divided into two categories: long, multidomain proteins (RelA, Rel and SpoT) and short, single-domain enzymes (small alarmone synthetases (SASs) and small alarmone hydrolases (SAHs)). The enzymatic activity of the long enzymes is regulated by interactions with molecular effectors, such as 'starved' ribosomes in the case of RelA, and the activity of short RSHs is regulated at the transcriptional level.
-
The regulatory role of (p)ppGpp in general metabolism is exerted by direct and indirect mechanisms. The direct mechanisms rely on the alarmone binding to and regulating a molecular target, whereas indirect mechanisms rely on changes in the concentrations of GTP and ATP, elicited by (p)ppGpp production.
-
The primary target of (p)ppGpp-mediated regulation is transcription. In Escherichia coli, this regulation relies on direct binding of the alarmone to the β′- and ω-subunits of RNA polymerase (RNAP), whereas in Bacillus subtilis, regulation is indirect and relies on changes in the concentration of the initiator nucleotide.
-
In addition to transcription, (p)ppGpp production regulates several other cellular processes, such as protein biosynthesis, replication, the acid stress response, polyphosphate metabolism, biosynthesis and uptake of nucleotides, and (via a direct interaction with RelA) the stringent response itself.
-
The effects of (p)ppGpp span a continuum from acute survival responses elicited by stresses such as nutrient deprivation or heat shock, mediated by high (p)ppGpp levels, to the 'housekeeping' role of basal (p)ppGpp levels in normal metabolic proesses, such as the production of amino acids and nucleotides.
-
The stringent response has a key role in bacterial virulence and persistence (the formation of antibiotic-tolerant cells). This has prompted the recent development of specific inhibitors of this process, which serve as a starting point for the future development of novel antivirulence compounds.
Abstract
The alarmones guanosine tetraphosphate and guanosine pentaphosphate (collectively referred to as (p)ppGpp) are involved in regulating growth and several different stress responses in bacteria. In recent years, substantial progress has been made in our understanding of the molecular mechanisms of (p)ppGpp metabolism and (p)ppGpp-mediated regulation. In this Review, we summarize these recent insights, with a focus on the molecular mechanisms governing the activity of the RelA/SpoT homologue (RSH) proteins, which are key players that regulate the cellular levels of (p)ppGpp. We also discuss the structural basis of transcriptional regulation by (p)ppGpp and the role of (p)ppGpp in GTP metabolism and in the emergence of bacterial persisters.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pesavento, C. & Hengge, R. Bacterial nucleotide-based second messengers. Curr. Opin. Microbiol. 12, 170–176 (2009).
Potrykus, K., Murphy, H., Philippe, N. & Cashel, M. ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol. 13, 563–575 (2011).
Kriel, A. et al. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol. Cell 48, 231–241 (2012).
Gaca, A. O. et al. Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. mBio 4, e00646–13 (2013).
Potrykus, K. & Cashel, M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51 (2008). A review that summarizes compelling evidence for the role of (p)ppGpp in growth rate control in E. coli.
Scott, M., Gunderson, C. W., Mateescu, E. M., Zhang, Z. & Hwa, T. Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–1102 (2010).
Gallant, J., Irr, J. & Cashel, M. The mechanism of amino acid control of guanylate and adenylate biosynthesis. J. Biol. Chem. 246, 5812–5816 (1971).
Hochstadt-Ozer, J. & Cashel, M. The regulation of purine utilization in bacteria. V. Inhibition of purine phosphoribosyltransferase activities and purine uptake in isolated membrane vesicles by guanosine tetra-phosphate. J. Biol. Chem. 247, 7067–7072 (1972).
Traxler, M. F. et al. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the 'feast to famine' gradient in Escherichia coli. Mol. Microbiol. 79, 830–845 (2011).
Atkinson, G. C., Tenson, T. & Hauryliuk, V. The RelA/SpoT Homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE 6, e23479 (2011). A comprehensive analysis of RSH evolution across the tree of life, providing a unifying nomenclature for this enzyme family.
Ito, D. et al. Enzymatic and molecular characterization of Arabidopsis ppGpp pyrophosphohydrolase, AtNUDX26. Biosci. Biotechnol. Biochem. 76, 2236–2241 (2012).
Ooga, T. et al. Degradation of ppGpp by Nudix pyrophosphatase modulates the transition of growth phase in the bacterium Thermus thermophilus. J. Biol. Chem. 284, 15549–15556 (2009).
Kraszewska, E. The plant Nudix hydrolase family. Acta Biochim. Pol. 55, 663–671 (2008).
Keasling, J. D., Bertsch, L. & Kornberg, A. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc. Natl Acad. Sci. USA 90, 7029–7033 (1993).
Hamel, E. & Cashel, M. Role of guanine nucleotides in protein synthesis. Elongation factor G and guanosine 5′-triphosphate, 3′-diphosphate. Proc. Natl Acad. Sci. USA 70, 3250–3254 (1973).
Dalebroux, Z. D., Svensson, S. L., Gaynor, E. C. & Swanson, M. S. ppGpp conjures bacterial virulence. Microbiol. Mol. Biol. Rev. 74, 171–199 (2010).
Geiger, T. et al. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 8, e1003016 (2012).
Poole, K. Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrobiol. Chemother. 67, 2069–2089 (2012).
Maisonneuve, E. & Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548 (2014). A detailed review of the molecular mechanisms of bacterial persistence, with a focus on the role of (p)ppGpp and the stringent response.
Cashel, M. & Gallant, J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221, 838–841 (1969).
Laffler, T. & Gallant, J. spoT, a new genetic locus involved in the stringent response in E. coli. Cell 1, 27–30 (1974).
Mechold, U., Murphy, H., Brown, L. & Cashel, M. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of RelSeq, the Rel/Spo enzyme from Streptococcus equisimilis. J. Bacteriol. 184, 2878–2888 (2002).
Seyfzadeh, M., Keener, J. & Nomura, M. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc. Natl Acad. Sci. USA 90, 11004–11008 (1993).
Vinella, D., Albrecht, C., Cashel, M. & D'Ari, R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol. Microbiol. 56, 958–970 (2005).
Xiao, H. et al. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266, 5980–5990 (1991).
Haseltine, W. A., Block, R., Gilbert, W. & Weber, K. MSI and MSII made on ribosome in idling step of protein synthesis. Nature 238, 381–384 (1972).
Gallant, J., Palmer, L. & Pao, C. C. Anomalous synthesis of ppGpp in growing cells. Cell 11, 181–185 (1977).
Shyp, V. et al. Positive allosteric feedback regulation of the stringent response enzyme RelA by its product. EMBO Rep. 13, 835–839 (2012). A study showing that ppGpp markedly increases the rate of its own synthesis by RelA through a positive allosteric feedback mechanism that had not been described previously for any enzyme.
Haseltine, W. A. & Block, R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl Acad. Sci. USA 70, 1564–1568 (1973). Biochemical investigations demonstrating tRNA- and ribosome-dependent activation of RelA-mediated (p)ppGpp synthesis.
Sprinzl, M. & Richter, D. Free 3′-OH group of the terminal adenosine of the tRNA molecule is essential for the synthesis in vitro of guanosine tetraphosphate and pentaphosphate in a ribosomal system from Escherichia coli. Eur. J. Biochem. 71, 171–176 (1976).
Payoe, R. & Fahlman, R. P. Dependence of RelA-mediated (p)ppGpp formation on tRNA identity. Biochemistry 50, 3075–3083 (2011).
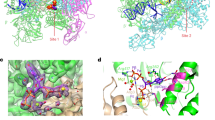
Wendrich, T. M., Blaha, G., Wilson, D. N., Marahiel, M. A. & Nierhaus, K. H. Dissection of the mechanism for the stringent factor RelA. Mol. Cell 10, 779–788 (2002). A report presenting the hopping model of RelA-mediated (p)ppGpp synthesis, in which (p)ppGpp synthesis proceeds following RelA dissociation from the ribosome.
Richter, D. Stringent factor from Escherichia coli directs ribosomal binding and release of uncharged tRNA. Proc. Natl Acad. Sci. USA 73, 707–711 (1976).
English, B. P. et al. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc. Natl Acad. Sci. USA 108, E365–E373 (2011). An in vivo single-molecule tracking analysis of RelA, the results of which suggest that ppGpp is synthesized by RelA when it is off, rather than on, the ribosome.
Agirrezabala, X. et al. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep. 14, 811–816 (2013). A cryo-EM analysis of RelA in complex with the ribosome.
Parker, J., Watson, R. J. & Friesen, J. D. A relaxed mutant with an altered ribosomal protein L11. Mol. Gen. Genet. 144, 111–114 (1976). A study demonstrating the crucial role of ribosomal protein L11 in the regulation of RelA.
Schmeing, T. M. et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–694 (2009).
Harms, J. M. et al. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell 30, 26–38 (2008).
An, G., Justesen, J., Watson, R. J. & Friesen, J. D. Cloning the spoT gene of Escherichia coli: identification of the spoT gene product. J. Bacteriol. 137, 1100–1110 (1979).
Raskin, D. M., Judson, N. & Mekalanos, J. J. Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae. Proc. Natl Acad. Sci. USA 104, 4636–4641 (2007).
Battesti, A. & Bouveret, E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 62, 1048–1063 (2006). A paper describing the molecular mechanism of SpoT regulation by ACP.
Yao, Z., Davis, R. M., Kishony, R., Kahne, D. & Ruiz, N. Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc. Natl Acad. Sci. USA 109, E2561–E2568 (2012).
Sy, J. In vitro degradation of guanosine 5′-diphosphate, 3′-diphosphate. Proc. Natl Acad. Sci. USA 74, 5529–5533 (1977).
Hara, A. & Sy, J. Guanosine 5′-triphosphate, 3′-diphosphate 5′-phosphohydrolase. Purification and substrate specificity. J. Biol. Chem. 258, 1678–1683 (1983).
Dahl, J. L. et al. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl Acad. Sci. USA 100, 10026–10031 (2003).
Primm, T. P. et al. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 182, 4889–4898 (2000).
Avarbock, D., Avarbock, A. & Rubin, H. Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA·ribosome·mRNA·RelMtb complex. Biochemistry 39, 11640–11648 (2000).
Scoarughi, G. L., Cimmino, C. & Donini, P. Helicobacter pylori: a eubacterium lacking the stringent response. J. Bacteriol. 181, 552–555 (1999).
Wells, D. H. & Gaynor, E. C. Helicobacter pylori initiates the stringent response upon nutrient and pH downshift. J. Bacteriol. 188, 3726–3729 (2006).
Park, S. A., Ko, A. & Lee, N. G. Stimulation of growth of the human gastric pathogen Helicobacter pylori by atmospheric level of oxygen under high carbon dioxide tension. BMC Microbiol. 11, 96 (2011).
Boutte, C. C. & Crosson, S. The complex logic of stringent response regulation in Caulobacter crescentus: starvation signalling in an oligotrophic environment. Mol. Microbiol. 80, 695–714 (2011).
Gaca, A. O., Abranches, J., Kajfasz, J. K. & Lemos, J. A. Global transcriptional analysis of the stringent response in Enterococcus faecalis. Microbiology 158, 1994–2004 (2012).
Harris, B. Z., Kaiser, D. & Singer, M. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 12, 1022–1035 (1998).
Strauch, E., Takano, E., Baylis, H. A. & Bibb, M. J. The stringent response in Streptomyces coelicolor A3(2). Mol. Microbiol. 5, 289–298 (1991).
Boutte, C. C. & Crosson, S. Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 21, 174–180 (2013).
Battesti, A. & Bouveret, E. Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J. Bacteriol. 191, 616–624 (2009).
Wout, P. et al. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol. 186, 5249–5257 (2004).
Persky, N. S., Ferullo, D. J., Cooper, D. L., Moore, H. R. & Lovett, S. T. The ObgE/CgtA GTPase influences the stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 73, 253–266 (2009).
Hogg, T., Mechold, U., Malke, H., Cashel, M. & Hilgenfeld, R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117, 57–68 (2004); erratum 117, 415 (2004). An X-ray-based structural analysis of a truncated Rel protein from S. dysgalactiae subsp. equisimilis , providing a structural model for the intramolecular regulation of Rel.
Paul, B. J., Berkmen, M. B. & Gourse, R. L. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl Acad. Sci. USA 102, 7823–7828 (2005).
Paul, B. J. et al. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118, 311–322 (2004). An investigation that identifies the transcriptional factor DksA as a regulator of RNAP, together with ppGpp.
Barker, M. M., Gaal, T., Josaitis, C. A. & Gourse, R. L. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305, 673–688 (2001).
Krasny, L. & Gourse, R. L. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 23, 4473–4483 (2004). A study showing that in B. subtilis , unlike in E. coli , the regulation of rRNA production by ppGpp occurs by an indirect mechanism that relies on an alteration in the concentration of initiator nucleotides, rather than on direct allosteric regulation of RNAP by ppGpp.
Artsimovitch, I. et al. Structural basis for transcription regulation by alarmone ppGpp. Cell 117, 299–310 (2004).
Vrentas, C. E. et al. Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J. Mol. Biol. 377, 551–564 (2008).
Mechold, U., Potrykus, K., Murphy, H., Murakami, K. S. & Cashel, M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 41, 6175–6189 (2013). Work demonstrating that ppGpp is a more potent effector than pppGpp in E. coli.
Zuo, Y., Wang, Y. & Steitz, T. A. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol. Cell 50, 430–436 (2013). A paper presenting the X-ray crystal structure of RNAP in complex with ppGpp.
Igarashi, K., Fujita, N. & Ishihama, A. Promoter selectivity of Escherichia coli RNA polymerase: omega factor is responsible for the ppGpp sensitivity. Nucleic Acids Res. 17, 8755–8765 (1989).
Vrentas, C. E., Gaal, T., Ross, W., Ebright, R. H. & Gourse, R. L. Response of RNA polymerase to ppGpp: requirement for the ω subunit and relief of this requirement by DksA. Genes Dev. 19, 2378–2387 (2005).
Ross, W., Vrentas, C. E., Sanchez-Vazquez, P., Gaal, T. & Gourse, R. L. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 50, 420–429 (2013).
Tagami, S. et al. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 468, 978–982 (2010).
Tagami, S., Sekine, S. I. & Yokoyama, S. A novel conformation of RNA polymerase sheds light on the mechanism of transcription. Transcription 2, 162–167 (2011).
Tare, P., Mallick, B. & Nagaraja, V. Co-evolution of specific amino acid in σ1.2 region and nucleotide base in the discriminator to act as sensors of small molecule effectors of transcription initiation in mycobacteria. Mol. Microbiol. 90, 569–583 (2013).
Kasai, K. et al. Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus. J. Bacteriol. 188, 7111–7122 (2006).
Lennon, C. W. et al. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev. 26, 2634–2646 (2012).
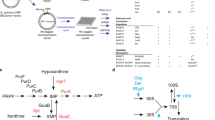
Jensen, K. F., Dandanell, G., Hove-Jensen, B. & Willemoës, M. in EcoSal — Escherichia coli and Salmonella: Cellular and Molecular Biology Ch. 3.6.2 (ed. Stewart, V.) (ASMscience, 2008).
Papakostas, K., Botou, M. & Frillingos, S. Functional identification of the hypoxanthine/guanine transporters YjcD and YgfQ and the adenine transporters PurP and YicO of Escherichia coli K-12. J. Biol. Chem. 288, 36827–36840 (2013).
Meng, L. M. & Nygaard, P. Identification of hypoxanthine and guanine as the co-repressors for the purine regulon genes of Escherichia coli. Mol. Microbiol. 4, 2187–2192 (1990).
Irr, J. & Gallant, J. The control of ribonucleic acid synthesis in Escherichia coli: II. Stringent control of energy metabolism. J. Biol. Chem. 244, 2233–2239 (1969).
Pao, C. C. & Dyess, B. T. Effect of unusual guanosine nucleotides on the activities of some Escherichia coli cellular enzymes. Biochim. Biophys. Acta 677, 358–362 (1981).
Nomura, Y. et al. Diversity in guanosine 3′,5′-bisdiphosphate (ppGpp) sensitivity among guanylate kinases of bacteria and plants. J. Biol. Chem. 289, 15631–15641 (2014).
Beaman, T. C. et al. Specificity and control of uptake of purines and other compounds in Bacillus subtilis. J. Bacteriol. 156, 1107–1117 (1983).
Kriel, A. et al. GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes. J. Bacteriol. 196, 189–201 (2014). A report highlighting the role of ppGpp and GTP levels in amino acid metabolism in B. subtilis.
Geiger, T. & Wolz, C. Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. Int. J. Med. Microbiol. 304, 150–155 (2014).
Bittner, A. N., Kriel, A. & Wang, J. D. Lowering GTP level increases survival of amino acid starvation but slows growth rate for Bacillus subtilis cells lacking (p)ppGpp. J. Bacteriol. 196, 2067–2076 (2014).
Bernardo, L. M. & Johansson, L. U., Skarfstad, E. & Shingler, V. σ54-promoter discrimination and regulation by ppGpp and DksA. J. Biol. Chem. 284, 828–838 (2009).
Lewis, K. Persister cells. Annu. Rev. Microbiol. 64, 357–372 (2010).
Levin, B. R., Concepcion-Acevedo, J. & Udekwu, K. I. Persistence: a copacetic and parsimonious hypothesis for the existence of non-inherited resistance to antibiotics. Curr. Opin. Microbiol. 21, 18–21 (2014).
Bigger, J. W. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244, 497–500 (1944).
Amato, S. M., Orman, M. A. & Brynildsen, M. P. Metabolic control of persister formation in Escherichia coli. Mol. Cell 50, 475–487 (2013).
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Korch, S. B., Henderson, T. A. & Hill, T. M. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50, 1199–1213 (2003).
Germain, E., Castro-Roa, D., Zenkin, N. & Gerdes, K. Molecular mechanism of bacterial persistence by HipA. Mol. Cell 52, 248–254 (2013).
Kaspy, I. et al. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nature Commun. 4, 3001 (2013).
Bokinsky, G. et al. HipA-triggered growth arrest and β-lactam tolerance in Escherichia coli are mediated by RelA-dependent ppGpp synthesis. J. Bacteriol. 195, 3173–3182 (2013).
Keren, I., Shah, D., Spoering, A., Kaldalu, N. & Lewis, K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186, 8172–8180 (2004).
Shah, D. et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6, 53 (2006).
Gerdes, K. & Maisonneuve, E. Bacterial persistence and toxin–antitoxin loci. Annu. Rev. Microbiol. 66, 103–123 (2012).
Hansen, S. et al. Regulation of the Escherichia coli HipBA toxin–antitoxin system by proteolysis. PLoS ONE 7, e39185 (2012).
Maisonneuve, E., Shakespeare, L. J., Jorgensen, M. G. & Gerdes, K. Bacterial persistence by RNA endonucleases. Proc. Natl Acad. Sci. USA 108, 13206–13211 (2011).
Maisonneuve, E., Castro-Camargo, M. & Gerdes, K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin–antitoxin activity. Cell 154, 1140–1150 (2013). A study that uncovers the mechanism behind the bacterial persistence phenotype induced by increased ppGpp levels.
Kuroda, A., Murphy, H., Cashel, M. & Kornberg, A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 272, 21240–21243 (1997).
Kuroda, A. et al. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293, 705–708 (2001).
Helaine, S. et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343, 204–208 (2014).
Claudi, B. et al. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell 158, 722–733 (2014).
Khakimova, M., Ahlgren, H. G., Harrison, J. J., English, A. M. & Nguyen, D. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J. Bacteriol. 195, 2011–2020 (2013).
Nguyen, D. et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986 (2011). A study demonstrating that the stringent response induces antibiotic tolerance by eliciting a specific cell survival programme rather than by slowing down bacterial growth.
Cashel, M. Regulation of bacterial ppGpp and pppGpp. Annu. Rev. Microbiol. 29, 301–318 (1975).
Dalebroux, Z. D. & Swanson, M. S. ppGpp: magic beyond RNA polymerase. Nature Rev. Microbiol. 10, 203–212 (2012). An excellent review summarizing the role of ppGpp in bacterial virulence.
Bibb, M. J. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8, 208–215 (2005).
Milon, P. et al. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc. Natl Acad. Sci. USA 103, 13962–13967 (2006).
Mitkevich, V. A. et al. Thermodynamic characterization of ppGpp binding to EF-G or IF2 and of initiator tRNA binding to free IF2 in the presence of GDP, GTP, or ppGpp. J. Mol. Biol. 402, 838–846 (2010).
Wang, J. D., Sanders, G. M. & Grossman, A. D. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128, 865–875 (2007).
Kanjee, U., Ogata, K. & Houry, W. A. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol. Microbiol. 85, 1029–1043 (2012).
Murdeshwar, M. S. & Chatterji, D. MS_RHII-RSD, a dual-function RNase HII-(p)ppGpp synthetase from Mycobacterium smegmatis. J. Bacteriol. 194, 4003–4014 (2012).
Nanamiya, H. et al. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67, 291–304 (2008). One of the first reports to identify a single-domain small alarmone synthetase enzyme.
Lemos, J. A., Lin, V. K., Nascimento, M. M., Abranches, J. & Burne, R. A. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol. Microbiol. 65, 1568–1581 (2007).
Cao, M. et al. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316, 443–457 (2002).
Sun, D. et al. A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nature Struct. Mol. Biol. 17, 1188–1194 (2010).
Thammana, P., Buerk, R. R. & Gordon, J. Absence of ppGpp production in synchronised Balb/C mouse 3T3 cells on isoleucine starvation. FEBS Lett. 68, 187–190 (1976).
Tozawa, Y. & Nomura, Y. Signalling by the global regulatory molecule ppGpp in bacteria and chloroplasts of land plants. Plant Biol. (Stuttg.) 13, 699–709 (2011).
O'Day, D. H. & Keszei, A. Signalling and sex in the social amoebozoans. Biol. Rev. Camb. Philos. Soc. 87, 313–329 (2012).
Rasko, D. A. & Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nature Rev. Drug Discov. 9, 117–128 (2010).
Wexselblatt, E. et al. Relacin, a novel antibacterial agent targeting the stringent response. PLoS Pathog. 8, e1002925 (2012).
Wexselblatt, E., Kaspy, I., Glaser, G., Katzhendler, J. & Yavin, E. Design, synthesis and structure–activity relationship of novel Relacin analogs as inhibitors of Rel proteins. Eur. J. Med. Chem. 70, 497–504 (2013).
de la Fuente-Nunez, C., Reffuveille, F., Haney, E. F., Straus, S. K. & Hancock, R. E. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 10, e1004152 (2014).
Ciccarelli, F. D. et al. Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283–1287 (2006).
Acknowledgements
The authors are grateful to V. Shingler for her comments on the manuscript and M. Valle for providing the cryo-EM images of RelA. This work was supported by the European Regional Development Fund through the Centre of Excellence in Chemical Biology (V.H. and T.T.); by the Estonian Science Foundation (grants ETF9012 and PUT37 (V.H.), and grant ETF9020 (G.C.A.)); by Umeå University, the Swedish Research Council, the Ragnar Söderberg Foundation and the Kempe Foundation (V.H.); by the US National Institutes of Health (grant GM087350 (K.S.M.)); and by a European Research Council Advanced Investigator Grant (294517; PERSIST) and a Novo Nordisk Foundation Laureate Research Grant (K.G.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- GTPases
-
Enzymes that bind and hydrolyse GTP to produce GDP.
- Translocation
-
A step in the elongation cycle of the ribosome in which the mRNA–tRNA complex advances on the ribosome by a distance of one codon.
- ThrRS, GTPase and SpoT domain
-
(TGS domain). A protein domain with a suggested ligand-binding function.
- Aspartokinase, chorismate mutase and TyrA domain
-
(ACT domain). A protein domain that is involved in ligand binding and that is often present in metabolic enzymes.
- A-site
-
The ribosome acceptor site, which accommodates tRNA and various protein factors during the translation cycle.
- Deacylated tRNAs
-
tRNAs that are uncharged, as they lack an aminoacyl group at the 3′-CCA end.
- Sarcin–ricin loop
-
(SRL). An essential structural element of the 23S ribosomal RNA; this loop interacts with various protein factors during translation.
- A/T state
-
A structural conformation of an aminoacyl-tRNA, in which the charged tRNA becomes distorted as it occupies the ribosomal A-site while interacting with elongation factor Tu.
- Elongation factor Tu
-
(EF-Tu). A GTPase that delivers aminoacyl-tRNAs to the ribosomal A-site during protein synthesis.
- Acyl carrier protein
-
(ACP). An essential component of the fatty acid synthesis pathway that stabilizes and transports the growing lipid chain. Specialized ACPs are also involved in other processes that require acyl transfer, such as the synthesis of polyketide antibiotics.
- σW regulon
-
An operon that is regulated by the transcription factor σW and that is induced in response to various stresses.
- Monophyletic
-
Pertaining to a clade in a phylogenetic tree: sharing a single common ancestor.
- Paraphyletic
-
Pertaining to a clade in a phylogenetic tree: sharing a single common ancestor but excluding some descendent sequences.
- Salvage pathway
-
A pathway in which nucleotides are synthesized from intermediates (nucleobases and ribonucleosides that are either the products of degradative processes within the cell or imported from the extracellular milieu), rather than de novo, from phosphoribosyl pyrophosphate as a starting compound.
- ppGpp0 strain
-
A bacterial strain that is unable to produce (p)ppGpp, as it lacks functional large and small RelA–SpoT homologues.
- Toxin–antitoxin module
-
(TA module). A bacterial system that involves a toxin and an antitoxin that neutralizes its cognate toxin (for type II TA systems). The toxins of these systems collectively inhibit several essential cellular functions, such as translation, DNA replication and cell wall synthesis.
- Lon protease
-
An ATP-dependent protease that degrades aberrant and short-lived polypeptides.
- Phenol-soluble modulins
-
(PSMs). A family of amphipathic α-helical peptides that have surfactant-like properties and multiple roles in staphylococcal virulence through their contribution to immune evasion and biofilm development.
- A-factor
-
A signalling molecule (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) that triggers secondary metabolism and morphogenesis in Streptomyces spp.
- Fruiting bodies
-
Multicellular aggregates of myxobacteria that form as a result of nutrient deprivation.
Rights and permissions
About this article
Cite this article
Hauryliuk, V., Atkinson, G., Murakami, K. et al. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13, 298–309 (2015). https://doi.org/10.1038/nrmicro3448
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro3448
This article is cited by
-
Phage Paride can kill dormant, antibiotic-tolerant cells of Pseudomonas aeruginosa by direct lytic replication
Nature Communications (2024)
-
Impacts of electrochemical disinfection on the viability and structure of the microbiome in secondary effluent water
Frontiers of Environmental Science & Engineering (2024)
-
Efficacy of outer membrane permeabilization in promoting aromatic isothiocyanates-mediated eradication of multidrug resistant Gram-negative bacteria and bacterial persisters
Folia Microbiologica (2024)
-
Glyphosate affects persistence and tolerance but not antibiotic resistance
BMC Microbiology (2023)
-
Structure of SpoT reveals evolutionary tuning of catalysis via conformational constraint
Nature Chemical Biology (2023)