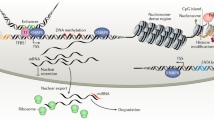
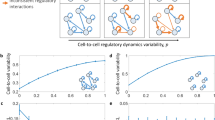
Abstract
Single-cell measurements and lineage-tracing experiments are revealing that phenotypic cell-to-cell variability is often the result of deterministic processes, despite the existence of intrinsic noise in molecular networks. In most cases, this determinism represents largely uncharacterized molecular regulatory mechanisms, which places the study of cell-to-cell variability in the realm of molecular cell biology. Further research in the field will be important to advance quantitative cell biology because it will provide new insights into the mechanisms by which cells coordinate their intracellular activities in the spatiotemporal context of the multicellular environment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Niepel, M., Spencer, S. & Sorger, P. Non-genetic cell-to-cell variability and the consequences for pharmacology. Curr. Opin. Chem. Biol. 13, 556–561 (2009).
Altschuler, S. J. & Wu, L. F. Cellular heterogeneity: do differences make a difference? Cell 141, 559–563 (2010).
Lee, T. & Covert, M. High-throughput, single-cell NF-κB dynamics. Curr. Opin. Genet. Dev. 20, 1–7 (2010).
Spiller, D., Wood, C., Rand, D. & White, M. Measurement of single-cell dynamics. Nature 465, 736–745 (2010).
Muzzey, D. & van Oudenaarden, A. Quantitative time-lapse fluorescence microscopy in single cells. Ann. Rev. Cell Dev. 25, 301–327 (2009).
Snijder, B. et al. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature 461, 520–523 (2009).
Singh, D. K. et al. Patterns of basal signaling heterogeneity can distinguish cellular populations with different drug sensitivities. Mol. Syst. Biol. 6, 369 (2010).
Cohen, A. A. et al. Dynamic proteomics of individual cancer cells in response to a drug. Science 322, 1511–1516 (2008).
Spencer, S., Gaudet, S., Albeck, J., Burke, J. & Sorger, P. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459, 428–432 (2009).
Brock, A., Chang, H. & Huang, S. Non-genetic heterogeneity — a mutation-independent driving force for the somatic evolution of tumours. Nature Rev. Genet. 10, 336–342 (2009).
Kumar, R., Kuniyasu, H., Bucana, C. D., Wilson, M. R. & Fidler, I. J. Spatial and temporal expression of angiogenic molecules during tumor growth and progression. Oncol. Res. 10, 301–311 (1998).
Hoek, K. S. et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 68, 650–656 (2008).
Roesch, A. et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 141, 583–594 (2010).
Ungrin, M., Joshi, C., Nica, A., Bauwens, C. & Zandstra, P. Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS ONE 3, e1565 (2008).
Discher, D. E., Mooney, D. J. & Zandstra, P. W. Growth factors, matrices, and forces combine and control stem cells. Science 324, 1673–1677 (2009).
Smith, Z., Nachman, I., Regev, A. & Meissner, A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nature Biotech. 28, 521–526 (2010).
Tay, S. et al. Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature 466, 267–271 (2010).
Zeng, L. et al. Decision making at a subcellular level determines the outcome of bacteriophage infection. Cell 141, 682–691 (2010).
St-Pierre, F. & Endy, D. Determination of cell fate selection during phage lambda infection. Proc. Natl Acad. Sci. USA 105, 20705–20710 (2008).
Colman-Lerner, A. et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature 437, 699–706 (2005).
Yu, R. et al. Negative feedback that improves information transmission in yeast signalling. Nature 456, 755–761 (2008).
Avery, S. V. Microbial cell individuality and the underlying sources of heterogeneity. Nature Rev. Microbiol. 4, 577–587 (2006).
Vlamakis, H., Aguilar, C., Losick, R. & Kolter, R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 22, 945–953 (2008).
Veening, J. et al. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc. Natl Acad. Sci. USA 105, 4393–4398 (2008).
Robert, L. et al. Pre-dispositions and epigenetic inheritance in the Escherichia coli lactose operon bistable switch. Mol. Syst. Biol. 6, 357 (2010).
Nachman, I., Regev, A. & Ramanathan, S. Dissecting timing variability in yeast meiosis. Cell 131, 544–556 (2007).
Maheshri, N. & O'Shea, E. K. Living with noisy genes: how cells function reliably with inherent variability in gene expression. Annu. Rev. Biophys. Biomol. Struct. 36, 413–434 (2007).
Raj, A. & van Oudenaarden, A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226 (2008).
Simpson, M. L. et al. Noise in biological circuits. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 1, 214–225 (2009).
Paulsson, J. Summing up the noise in gene networks. Nature 427, 415–418 (2004).
Losick, R. & Desplan, C. Stochasticity and cell fate. Science 320, 65–68 (2008).
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002).
Ozbudak, E., Thattai, M., Kurtser, I., Grossman, A. & van Oudenaarden, A. Regulation of noise in the expression of a single gene. Nature genet. 31, 69–73 (2002).
Raj, A. & van Oudenaarden, A. Single-molecule approaches to stochastic gene expression. Ann. Rev. Biophys. 38, 255–270 (2009).
Ben-Jacob, E. & Schultz, D. Bacteria determine fate by playing dice with controlled odds. Proc. Natl Acad. Sci. USA 107, 13197–13198 (2010).
Eldar, A. & Elowitz, M. Functional roles for noise in genetic circuits. Nature 467, 167–173 (2010).
Taniguchi, Y. et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538 (2010).
Newman, J. et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441, 840–846 (2006).
Shahrezaei, V., Ollivier, J. & Swain, P. Colored extrinsic fluctuations and stochastic gene expression. Mol. Syst. Biol. 4, 196 (2008).
Volfson, D. et al. Origins of extrinsic variability in eukaryotic gene expression. Nature 439, 861–864 (2005).
Noise (entry 11a). OED online[online], (2010).
Gygi, S., Rochon, Y., Franza, B. & Aebersold, R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720 (1999).
Dehmelt, L. & Bastiaens, P. Spatial organization of intracellular communication: insights from imaging. Nature Rev. Mol. Cell Biol. 11, 440–452 (2010).
Scita, G. & Di Fiore, P. The endocytic matrix. Nature 463, 464–473 (2010).
Hunter, T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. cell 28, 730–738 (2007).
Rual, J. et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature 437, 1173–1178 (2005).
Tong, A. et al. Global mapping of the yeast genetic interaction network. Science 303, 808–813 (2004).
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Neumann, B., Walter, T. & Jean-Karim, H. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721–727 (2010).
Collinet, C. et al. Systems survey of endocytosis by multiparametric image analysis. Nature 464, 243–249 (2010).
Stelling, J., Sauer, U., Szallasi, Z., Doyle, F. J. 3rd & Doyle, J. Robustness of cellular functions. Cell 118, 675–685 (2004).
Lestas, I., Vinnicombe, G. & Paulsson, J. Fundamental limits on the suppression of molecular fluctuations. Nature 467, 174–178 (2010).
Sopko, R. et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol. cell 21, 319–330 (2006).
Isalan, M. et al. Evolvability and hierarchy in rewired bacterial gene networks. Nature 452, 840–845 (2008).
Delbrück, M. Statistical fluctuations in autocatalytic reactions. J. Chem. Phys. 8, 120–124 (1940).
Delbrück, M. The burst size distribution in the growth of bacterial viruses (bacteriophages). J. Bacteriol. 50, 131–135 (1945).
Novick, A. & Weiner, M. Enzyme induction as an all-or-none phenomenon. Proc. Natl Acad. Sci. USA 43, 553–566 (1957).
Gillespie, D. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361 (1977).
Arkin, A., Ross, J. & McAdams, H. H. Stochastic kinetic analysis of developmental pathway bifurcation in phage λ-infected Escherichia coli cells. Genetics 149, 1633–1648 (1998).
Herskowitz, I. & Hagen, D. The lysis-lysogeny decision of phage lambda: explicit programming and responsiveness. Annu. Rev. Genet. 14, 399–445 (1980).
Spudich, J. & Koshland, D. Non-genetic individuality: chance in the single cell. Nature 262, 467–471 (1976).
Eagle, H. & Levine, E. Growth regulatory effects of cellular interaction. Nature 213, 1102–1106 (1967).
Castor, L. Flattening, movement and control of division of epithelial-like cells. J. Cell. Physiol. 75, 57–64 (1970).
Colman-Lerner, A. et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature 437, 699–706 (2005).
Malleshaiah, M., Shahrezaei, V., Swain, P. & Michnick, S. The scaffold protein Ste5 directly controls a switch-like mating decision in yeast. Nature 465, 101–105 (2010).
Sigal, A. et al. Variability and memory of protein levels in human cells. Nature 444, 643–646 (2006).
Keren, K. et al. Mechanism of shape determination in motile cells. Nature 453, 475–480 (2008).
Schauer, K. et al. Probabilistic density maps to study global endomembrane organization. Nature Meth. 7, 560–566 (2010).
Ben-Jacob, E. Learning from bacteria about natural information processing. Ann. N. Y. Acad. Sci. 1178, 78–90 (2009).
Shapiro, J. Thinking about bacterial populations as multicellular organisms. Ann. Rev. Microbiol. 52, 81–104 (1998).
Waters, C. & Bassler, B. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346 (2005).
Blango, M. & Mulvey, M. Bacterial landlines: contact-dependent signaling in bacterial populations. Curr. Opin. Microbiol. 12, 177–181 (2009).
Bischofs, I., Hug, J., Liu, A., Wolf, D. & Arkin, A. Complexity in bacterial cell–cell communication: Quorum signal integration and subpopulation signaling in the Bacillus subtilis phosphorelay. Proc. Natl Acad. Sci. USA 106, 6459–6464 (2009).
Schultz, D., Wolynes, P. G., Ben Jacob, E. & Onuchic, J. N. Deciding fate in adverse times: sporulation and competence in Bacillus subtilis. Proc. Natl Acad. Sci. USA 106, 21027–21034 (2009).
Dubnau, D. & Losick, R. Bistability in bacteria. Mol. Microbiol. 61, 564–572 (2006).
Suel, G. M., Kulkarni, R. P., Dworkin, J., Garcia-Ojalvo, J. & Elowitz, M. B. Tunability and noise dependence in differentiation dynamics. Science 315, 1716–1719 (2007).
Wolf, D. et al. Memory in microbes: quantifying history-dependent behavior in a bacterium. PLoS ONE 3, e1700 (2008).
López, D. & Kolter, R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol. Rev. 34, 134–149 (2009).
Slack, M. D., Martinez, E. D., Wu, L. F. & Altschuler, S. J. Characterizing heterogeneous cellular responses to perturbations. Proc. Natl Acad. Sci. USA 105, 19306–19311 (2008).
Peng, S., Maihle, N. J. & Huang, Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene 29, 2153–2159 (2010).
Zernicka-Goetz, M., Morris, S. & Bruce, A. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nature Rev. Genet. 10, 467–477 (2009).
Keller, P., Schmidt, A., Wittbrodt, J. & Stelzer, E. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069 (2008).
Stockholm, D. et al. The origin of phenotypic heterogeneity in a clonal cell population in vitro. PLoS ONE 2, 394 (2007).
Chang, H., Hemberg, M., Barahona, M., Ingber, D. & Huang, S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 453, 544–547 (2008).
Sigismund, S. et al. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl Acad. Sci. USA 102, 2760–2765 (2005).
Mayor, S. & Pagano, R. Pathways of clathrin-independent endocytosis. Nature Rev. Mol. Cell Biol. 8, 603–612 (2007).
Altschuler, S., Angenent, S., Wang, Y. & Wu, L. On the spontaneous emergence of cell polarity. Nature 454, 886–889 (2008).
Sachs, K., Perez, O., Pe'er, D., Lauffenburger, D. A. & Nolan, G. P. Causal protein-signaling networks derived from multiparameter single-cell data. Science 308, 523–529 (2005).
Dunlop, M. J., Cox, R. S. 3rd, Levine, J. H., Murray, R. M. & Elowitz, M. B. Regulatory activity revealed by dynamic correlations in gene expression noise. Nature Genet. 40, 1493–1498 (2008).
Grecco, H. et al. In situ analysis of tyrosine phosphorylation networks by FLIM on cell arrays. Nature Meth. 7, 467–472 (2010).
Perfetto, S., Chattopadhyay, P. & Roederer, M. Seventeen-colour flow cytometry: unravelling the immune system. Nature Rev. Immunol. 4, 648–655 (2004).
Carpenter, A. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Ramo, P., Sacher, R., Snijder, B., Begemann, B. & Pelkmans, L. CellClassifier: supervised learning of cellular phenotypes. Bioinformatics 25, 3028–3030 (2009).
Bakal, C., Aach, J., Church, G. & Perrimon, N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science 316, 1753–1756 (2007).
Janes, K., Wang, C., Holmberg, K., Cabral, K. & Brugge, J. Identifying single-cell molecular programs by stochastic profiling. Nature Meth. 7, 311–317 (2010).
Damm, E. M. & Pelkmans, L. Systems biology of virus entry in mammalian cells. Cell. Microbiol. 8, 1219–1227 (2006).
Marsh, M. & Helenius, A. Virus entry: open sesame. Cell 124, 729–740 (2006).
Pelkmans, L. et al. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436, 78–86 (2005).
Mangan, S. & Alon, U. Structure and function of the feed-forward loop network motif. Proc. Natl Acad. Sci. USA 100, 11980–11985 (2003).
Miller, M. B. & Bassler, B. L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199 (2001).
Acknowledgements
We thank all members of the laboratory for stimulating discussions. Research in the L.P. laboratory is funded by the Swiss National Science Foundation, SystemsX.ch, the European Union; the Swiss Federal Institute of Technology (ETH) Zürich and the University of Zürich.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Related links
Rights and permissions
About this article
Cite this article
Snijder, B., Pelkmans, L. Origins of regulated cell-to-cell variability. Nat Rev Mol Cell Biol 12, 119–125 (2011). https://doi.org/10.1038/nrm3044
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3044
This article is cited by
-
The population context is a driver of the heterogeneous response of epithelial cells to interferons
Molecular Systems Biology (2024)
-
Measuring cell-to-cell expression variability in single-cell RNA-sequencing data: a comparative analysis and applications to B cell aging
Genome Biology (2023)
-
Extended methods for spatial cell classification with DBSCAN-CellX
Scientific Reports (2023)
-
MIL-CELL: a tool for multi-scale simulation of yeast replication and prion transmission
European Biophysics Journal (2023)
-
Einzelzellmikroskopie im Hochdurchsatz auf Mikrostrukturen
BIOspektrum (2022)