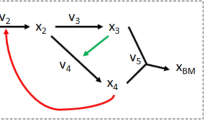
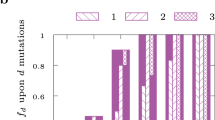
Key Points
-
Understanding the genotype–phenotype relationship of a cell has been a long-standing goal in biology. The availability of genome sequencing and of other high-throughput 'omic' data sets provides an opportunity to parameterize various statistical and mechanistic models.
-
Genome-scale metabolic network reconstructions contain comprehensively curated and systematized information on the cellular metabolism of an organism. These networks can be converted into a mathematical format that is amenable to constraint-based modelling.
-
Biological phenotype is constrained by the complex relationships between the genotype of a cell, its environment and physico-chemical laws. Rather than deriving a single solution to a problem, the philosophy of constraint-based modelling is to impose known constraints on physiological function to study the set of possible solutions.
-
Constraint-based modelling has a 30-year history, which can be separated into four phases — from initial theoretical interest and conception to maturing to predictive biological practice. In the current fourth phase, there is a critical mass of studies that combine high-throughput data and constraint-based models (CBMs) to answer relevant biological questions in a prospective manner.
-
Key successes in this field of research include deriving underlying principles for both optimal flux states and protein evolution; experimentally discovering cancer drug targets and antibiotics; and designing organisms that overproduce metabolic precursors of commodity chemicals.
-
Integrated modelling efforts combine CBMs with alternative modelling frameworks that are more amenable to capture other cellular processes in order to expand predictive scope outside metabolism. Expansions for other cellular features — such as transcription and translation machinery, transcriptional regulation and structural protein properties — have been constructed.
-
The formulation of high-dimensional models is enabled by the availability of genome-sequences (that is, a parts list), high-throughput omic data (that is, a functional readout) and CBMs (that is, a mechanistic framework to reconcile network topology, primary literature and omic data). These models can be used to quantitatively study the genotype–phenotype relationship for cellular metabolism, which will lead to a new generation of genome-scale science.
Abstract
The prediction of cellular function from a genotype is a fundamental goal in biology. For metabolism, constraint-based modelling methods systematize biochemical, genetic and genomic knowledge into a mathematical framework that enables a mechanistic description of metabolic physiology. The use of constraint-based approaches has evolved over ~30 years, and an increasing number of studies have recently combined models with high-throughput data sets for prospective experimentation. These studies have led to validation of increasingly important and relevant biological predictions. As reviewed here, these recent successes have tangible implications in the fields of microbial evolution, interaction networks, genetic engineering and drug discovery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Feist, A. M., Herrgard, M. J., Thiele, I., Reed, J. L. & Palsson, B. O. Reconstruction of biochemical networks in microorganisms. Nature Rev. Microbiol. 7, 129–143 (2009). This is a review on constructing and validating a genome-scale metabolic network.
Thiele, I. & Palsson, B. O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nature Protoc. 5, 93–121 (2010).
Lewis, N. E., Nagarajan, H. & Palsson, B. O. Constraining the metabolic genotype–phenotype relationship using a phylogeny of in silico methods. Nature Rev. Microbiol. 10, 291–305 (2012). This is a thorough review of the various constraint-based modelling methodologies.
Zhuang, K. et al. Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments. ISME J. 5, 305–316 (2011).
Klitgord, N. & Segre, D. Environments that induce synthetic microbial ecosystems. PLoS Comput. Biol. 6, e1001002 (2010).
Bordbar, A. et al. A multi-tissue type genome-scale metabolic network for analysis of whole-body systems physiology. BMC Syst. Biol. 5, 180 (2011).
Bordbar, A., Lewis, N. E., Schellenberger, J., Palsson, B. O. & Jamshidi, N. Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions. Mol. Syst. Biol. 6, 422 (2010).
Lewis, N. E. et al. Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nature Biotech. 28, 1279–1285 (2010).
Papin, J. A. & Palsson, B. O. The JAK–STAT signaling network in the human B-cell: an extreme signaling pathway analysis. Biophys. J. 87, 37–46 (2004).
Li, F., Thiele, I., Jamshidi, N. & Palsson, B. O. Identification of potential pathway mediation targets in Toll-like receptor signaling. PLoS Comput. Biol. 5, e1000292 (2009).
Gianchandani, E. P., Joyce, A. R., Palsson, B. O. & Papin, J. A. Functional states of the genome-scale Escherichia coli transcriptional regulatory system. PLoS Comput. Biol. 5, e1000403 (2009).
Thiele, I., Jamshidi, N., Fleming, R. M. & Palsson, B. O. Genome-scale reconstruction of Escherichia coli's transcriptional and translational machinery: a knowledge base, its mathematical formulation, and its functional characterization. PLoS Comput. Biol. 5, e1000312 (2009).
Fell, D. A. & Small, J. R. Fat synthesis in adipose tissue. An examination of stoichiometric constraints. Biochem. J. 238, 781–786 (1986).
Majewski, R. A. & Domach, M. M. Simple constrained optimization view of acetate overflow in E. coli. Biotechnol. Bioeng. 35, 732–738 (1990).
Savinell, J. M. & Palsson, B. O. Optimal selection of metabolic fluxes for in vivo measurement. II. Application to Escherichia coli and hybridoma cell metabolism. J. Theor. Biol. 155, 215–242 (1992).
Varma, A. & Palsson, B. O. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl. Environ. Microbiol. 60, 3724–3731 (1994).
Schuster, S. & Hilgetag, C. On elementary flux modes in biochemical reaction systems at steady state. J. Biol. Systems 2, 165–182 (1994).
Schilling, C. H., Letscher, D. & Palsson, B. O. Theory for the systemic definition of metabolic pathways and their use in interpreting metabolic function from a pathway-oriented perspective. J. Theor. Biol. 203, 229–248 (2000).
Clarke, B. L. in Advances in Chemical Physics Vol. 43 (eds. Prigogine, I. & Rice, S. A.) 1–215 (Wiley, 1980).
Dandekar, T., Schuster, S., Snel, B., Huynen, M. & Bork, P. Pathway alignment: application to the comparative analysis of glycolytic enzymes. Biochem. J. 343, 115–124 (1999).
Liao, J. C., Hou, S. Y. & Chao, Y. P. Pathway analysis, engineering and physiological considerations for redirecting central metabolism. Biotechnol. Bioeng. 52, 129–140 (1996).
Fleischmann, R. D. et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512 (1995).
Edwards, J. S. & Palsson, B. O. Systems properties of the Haemophilus influenzae Rd metabolic genotype. J. Biol. Chem. 274, 17410–17416 (1999).
Edwards, J. S., Ibarra, R. U. & Palsson, B. O. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nature Biotech. 19, 125–130 (2001).
Segre, D., Vitkup, D. & Church, G. M. Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl Acad. Sci. USA 99, 15112–15117 (2002).
Stelling, J., Klamt, S., Bettenbrock, K., Schuster, S. & Gilles, E. D. Metabolic network structure determines key aspects of functionality and regulation. Nature 420, 190–193 (2002).
Ibarra, R. U., Edwards, J. S. & Palsson, B. O. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420, 186–189 (2002).
Almaas, E., Kovacs, B., Vicsek, T., Oltvai, Z. N. & Barabasi, A. L. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature 427, 839–843 (2004).
Papp, B., Pal, C. & Hurst, L. D. Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature 429, 661–664 (2004).
Pal, C., Papp, B. & Lercher, M. J. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nature Genet. 37, 1372–1375 (2005).
Hyduke, D. R., Lewis, N. E. & Palsson, B. O. Analysis of omics data with genome-scale models of metabolism. Mol. Biosyst 9, 167–174 (2013). This is a review of techniques to integrate omic data with CBMs.
Patil, K. R. & Nielsen, J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc. Natl Acad. Sci. USA 102, 2685–2689 (2005).
Kharchenko, P., Church, G. M. & Vitkup, D. Expression dynamics of a cellular metabolic network. Mol Syst Biol 1, 2005.0016 (2005).
Shlomi, T., Cabili, M. N., Herrgard, M. J., Palsson, B. O. & Ruppin, E. Network-based prediction of human tissue-specific metabolism. Nature Biotech. 26, 1003–1010 (2008).
Becker, S. A. & Palsson, B. O. Context-specific metabolic networks are consistent with experiments. PLoS Comput. Biol. 4, e1000082 (2008).
Carlson, R. & Srienc, F. Fundamental Escherichia coli biochemical pathways for biomass and energy production: creation of overall flux states. Biotechnol. Bioeng. 86, 149–162 (2004).
Carlson, R. & Srienc, F. Fundamental Escherichia coli biochemical pathways for biomass and energy production: identification of reactions. Biotechnol. Bioeng. 85, 1–19 (2004).
Harcombe, W. R., Delaney, N. F., Leiby, N., Klitgord, N. & Marx, C. J. The ability of flux balance analysis to predict evolution of central metabolism scales with the initial distance to the optimum. PLoS Comput. Biol. 9, e1003091 (2013).
Schuetz, R., Kuepfer, L. & Sauer, U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol Syst Biol. 3, 119 (2007).
Molenaar, D., van Berlo, R., de Ridder, D. & Teusink, B. Shifts in growth strategies reflect tradeoffs in cellular economics. Mol. Syst. Biol. 5, 323 (2009).
Schuetz, R., Zamboni, N., Zampieri, M., Heinemann, M. & Sauer, U. Multidimensional optimality of microbial metabolism. Science 336, 601–604 (2012).
Lewis, N. E. et al. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol. Syst. Biol. 6, 390 (2010).
Khersonsky, O. & Tawfik, D. S. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505 (2010).
Nam, H. et al. Network context and selection in the evolution to enzyme specificity. Science 337, 1101–1104 (2012).
Feist, A. M. et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol 3, 121 (2007).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2, 2006.0008 (2006).
Scheer, M. et al. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 39, D670–D676 (2011).
Lobel, L., Sigal, N., Borovok, I., Ruppin, E. & Herskovits, A. A. Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence. PLoS Genet. 8, e1002887 (2012).
Costanzo, M. et al. The genetic landscape of a cell. Science 327, 425–431 (2010).
Uetz, P. et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 (2000).
Gama-Castro, S. et al. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units). Nucleic Acids Res. 39, D98–D105 (2011).
Segre, D., DeLuna, A., Church, G. M. & Kishnoy, R. Modular epistasis in yeast metabolism. Nature Genet. 37, 77–83 (2005).
Harrison, R., Papp, B., Pal, C., Oliver, S. G. & Delneri, D. Plasticity of genetic interactions in metabolic networks of yeast. Proc. Natl Acad. Sci. USA 104, 2307–2312 (2007).
He, X., Qian, W., Wang, Z., Li, Y. & Zhang, J. Prevalent positive epistasis in Escherichia coli and Saccharomyces cerevisiae metabolic networks. Nature Genet. 42, 272–276 (2010).
Szappanos, B. et al. An integrated approach to characterize genetic interaction networks in yeast metabolism. Nature Genet. 43, 656–662 (2011).
Mo, M. L., Palsson, B. O. & Herrgard, M. J. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst. Biol. 3, 37 (2009).
Wessely, F. et al. Optimal regulatory strategies for metabolic pathways in Escherichia coli depending on protein costs. Mol. Syst. Biol. 7, 515 (2011).
Notebaart, R. A., Teusink, B., Siezen, R. J. & Papp, B. Co-regulation of metabolic genes is better explained by flux coupling than by network distance. PLoS Comput. Biol. 4, e26 (2008).
Kaleta, C., de Figueiredo, L. F. & Schuster, S. Can the whole be less than the sum of its parts? Pathway analysis in genome-scale metabolic networks using elementary flux patterns. Genome Res. 19, 1872–1883 (2009).
Faith, J. J. et al. Many Microbe Microarrays Database: uniformly normalized Affymetrix compendia with structured experimental metadata. Nucleic Acids Res. 36, D866–D870 (2008).
Orth, J. D. & Palsson, B. O. Systematizing the generation of missing metabolic knowledge. Biotechnol. Bioeng. 107, 403–412 (2010). This is a review on techniques and applications of CBMs for a targeted expansion of biochemical knowledge.
Reed, J. L. et al. Systems approach to refining genome annotation. Proc. Natl Acad. Sci. USA 103, 17480–17484 (2006).
Duarte, N. C. et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl Acad. Sci. USA 104, 1777–1782 (2007).
Rolfsson, O., Paglia, G., Magnusdottir, M., Palsson, B. O. & Thiele, I. Inferring the metabolism of human orphan metabolites from their metabolic network context affirms human gluconokinase activity. Biochem. J. 449, 427–435 (2013).
Kanehisa, M., Goto, S., Sato, Y., Furumichi, M. & Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–D114 (2012).
Nakahigashi, K. et al. Systematic phenome analysis of Escherichia coli multiple-knockout mutants reveals hidden reactions in central carbon metabolism. Mol. Syst. Biol. 5, 306 (2009).
Lee, S. Y., Lee, D. Y. & Kim, T. Y. Systems biotechnology for strain improvement. Trends Biotechnol. 23, 349–358 (2005).
Park, J. H. & Lee, S. Y. Towards systems metabolic engineering of microorganisms for amino acid production. Curr. Opin. Biotechnol. 19, 454–460 (2008). This is a review of using systems biology methodologies for metabolic engineering applications.
Caspeta, L. & Nielsen, J. Economic and environmental impacts of microbial biodiesel. Nature Biotech. 31, 789–793 (2013).
Yim, H. et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nature Chem. Biol. 7, 445–452 (2011).
Hatzimanikatis, V. et al. Exploring the diversity of complex metabolic networks. Bioinformatics 21, 1603–1609 (2005).
Constantinou, L. & Gani, R. New group-contribution method for estimating properties of pure compounds. AIChE J. 40, 1697–1710 (1994).
Khatri, P., Sirota, M. & Butte, A. J. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput. Biol. 8, e1002375 (2012).
Burgard, A. P., Pharkya, P. & Maranas, C. D. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 84, 647–657 (2003).
Oberhardt, M. A., Yizhak, K. & Ruppin, E. Metabolically re-modeling the drug pipeline. Curr. Opin. Pharmacol. 13, 778–785 (2013). This is a review on using constraint-based modelling for drug discovery.
Hsu, P. P. & Sabatini, D. M. Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707 (2008).
Folger, O. et al. Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 7, 501 (2011).
Frezza, C. et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature 477, 225–228 (2011).
Jerby, L., Shlomi, T. & Ruppin, E. Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol. Syst. Biol. 6, 401 (2010).
Kim, P. J. et al. Metabolite essentiality elucidates robustness of Escherichia coli metabolism. Proc. Natl Acad. Sci. USA 104, 13638–13642 (2007).
Kim, H. U. et al. Integrative genome-scale metabolic analysis of Vibrio vulnificus for drug targeting and discovery. Mol. Syst. Biol. 7, 460 (2011).
Brynildsen, M. P., Winkler, J. A., Spina, C. S., MacDonald, I. C. & Collins, J. J. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nature Biotech. 31, 160–165 (2013).
Lerman, J. A. et al. In silico method for modelling metabolism and gene product expression at genome scale. Nature Commun. 3, 929 (2012).
Zhang, Y. et al. Three-dimensional structural view of the central metabolic network of Thermotoga maritima. Science 325, 1544–1549 (2009).
Thiele, I., Fleming, R. M., Bordbar, A., Schellenberger, J. & Palsson, B. O. Functional characterization of alternate optimal solutions of Escherichia coli's transcriptional and translational machinery. Biophys. J. 98, 2072–2081 (2010).
Pramanik, J. & Keasling, J. D. Effect of Escherichia coli biomass composition on central metabolic fluxes predicted by a stoichiometric model. Biotechnol. Bioeng. 60, 230–238 (1998).
Rodionova, I. A. et al. Diversity and versatility of the Thermotoga maritima sugar kinome. J. Bacteriol. 194, 5552–5563 (2012).
O'Brien, E. J., Lerman, J. A., Chang, R. L., Hyduke, D. R. & Palsson, B. O. Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction. Mol. Syst. Biol. 9, 693 (2013).
Chandrasekaran, S. & Price, N. D. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 107, 17845–17850 (2010).
Covert, M. W., Knight, E. M., Reed, J. L., Herrgard, M. J. & Palsson, B. O. Integrating high-throughput and computational data elucidates bacterial networks. Nature 429, 92–96 (2004).
Chang, R. L. et al. Structural systems biology evaluation of metabolic thermotolerance in Escherichia coli. Science 340, 1220–1223 (2013).
Gu, J. & Bourne, P. E. Structural bioinformatics (Wiley-Blackwell, 2009).
Marr, A. G. & Ingraham, J. L. Effect of temperature on the composition of fatty acids in Escherichia coli. J. Bacteriol. 84, 1260–1267 (1962).
Tenaillon, O. et al. The molecular diversity of adaptive convergence. Science 335, 457–461 (2012).
Mörters, P., Peres, Y., Schramm, O. & Werner, W. Brownian motion (Cambridge Univ. Press, 2010).
Karr, J. R. et al. A whole-cell computational model predicts phenotype from genotype. Cell 150, 389–401 (2012).
Thiele, I. et al. A community-driven global reconstruction of human metabolism. Nature Biotech. 31, 419–425 (2013).
Borenstein, E. Computational systems biology and in silico modeling of the human microbiome. Brief Bioinform. 13, 769–780 (2012).
Levy, R. & Borenstein, E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc. Natl Acad. Sci. USA 110, 12804–12809 (2013).
Atkinson, D. E. The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry 7, 4030–4034 (1968).
Weisz, P. B. Diffusion and chemical transformation. Science 179, 433–440 (1973).
Reed, J. L. Shrinking the metabolic solution space using experimental datasets. PLoS Comput. Biol. 8, e1002662 (2012). This is a review of the potential constraints that have been placed on CBMs.
Colijn, C. et al. Interpreting expression data with metabolic flux models: predicting Mycobacterium tuberculosis mycolic acid production. PLoS Comput. Biol. 5, e1000489 (2009).
Orth, J. D., Thiele, I. & Palsson, B. O. What is flux balance analysis? Nature Biotech. 28, 245–248 (2010). This paper presents a primer on the theory, applications and software toolboxes for FBA.
Mahadevan, R. & Schilling, C. H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 5, 264–276 (2003).
Wilkinson, D. J. Stochastic modelling for quantitative description of heterogeneous biological systems. Nature Rev. Genet. 10, 122–133 (2009).
Steuer, R. Computational approaches to the topology, stability and dynamics of metabolic networks. Phytochemistry 68, 2139–2151 (2007).
de Jong, H. Modeling and simulation of genetic regulatory systems: a literature review. J. Comput. Biol. 9, 67–103 (2002).
Friedman, N., Linial, M., Nachman, I. & Pe'er, D. Using Bayesian networks to analyze expression data. J. Computat. Biol. 7, 601–620 (2000).
Stephens, M. & Balding, D. J. Bayesian statistical methods for genetic association studies. Nature Rev. Genet. 10, 681–690 (2009).
Ideker, T. & Krogan, N. J. Differential network biology. Mol. Syst. Biol. 8, 565 (2012).
Califano, A., Butte, A. J., Friend, S., Ideker, T. & Schadt, E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nature Genet. 44, 841–847 (2012).
Acknowledgements
The authors thank D. Zielinski, J. Lerman, N. E. Lewis and H. Nagarajan for their criticisms and comments on the manuscript. This work was supported by the US National Institutes of Health grants GM068837 and GM057089, and by the Novo Nordisk Foundation. Z.A.K. is supported through the US National Science Foundation Graduate Research Fellowship (DGE-1144086).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Metabolite overflows
-
Biological phenomena whereby the rate of substrate use by a cell for growth is lower than the rates of uptake and conversion of the substrate, which results in production of side metabolites (for example, acetate in Escherichia coli).
- Metabolic fluxes
-
The rates of turnover or movement of metabolites through a reaction or a pathway.
- Objective functions
-
The particular variables, or metabolic reactions, that are being maximized or minimized for by the linear programme. In flux-balance analysis, the objective function is often a pseudoreaction for biomass generation that represents cellular growth.
- Metabolic pathways
-
In the context of this Review, sets of pathways that are calculated by metabolic network-based pathway analysis tools such as Extreme Pathways and Elementary Flux Modes.
- Metabolic engineering
-
The practice of improving cellular production of target compounds of interest by modifying and optimizing genetic, regulatory and environmental parameters of cellular metabolism.
- Genome-scale models
-
The formulation, using mathematical models, of genome-scale metabolic network reconstructions. They are synonymous with constraint-based models in the context of this Review.
- Pathway enrichment analysis
-
A high-throughput data analysis technique to understand more global changes in an experiment by grouping individual measurements of biological components (for example, genes and proteins) into a context that is based on various pathway databases (for example, Kyoto Encyclopedia of Genes and Genomes, BioCyc and Gene Ontology).
- Metabolic flux analysis
-
An experimental approach to identify metabolic fluxes using isotopically labelled metabolites and computational software that reconciles experimental data with network topology.
- Flux distributions
-
Sets of calculated flux values for all reactions in a constraint-based model.
- Pareto surface
-
The space that is formed when multiple objective functions are modelled at once; it represents a set of optimal solutions, in which increasing the value of one of the objectives results in a trade-off with other objective values.
- Central carbon metabolism
-
The metabolic pathways and reactions that convert sugars into the metabolic precursors that are required for growth. It is typically comprised of glycolysis, pentose phosphate pathways and the tricarboxylic acid cycle.
- Solution space
-
The range of all feasible values for variables in a constraint-based model, which represents all potential metabolic reaction flux distributions on the basis of the given constraints.
- Machine learning method
-
A method that applies statistical methods to discover generalizable rules and patterns in complex data sets.
- Gap-filling
-
Pertaining to a procedure for targeted expansion of metabolic knowledge, whereby prospective experiments are designed on the basis of discrepancies in experimental data and model predictions.
- Auxotrophies
-
Metabolic limitations that impair the ability of a cell or organism to synthesize a particular metabolite that is essential for growth, which force the cell or organism to rely on an exogenous source of the nutrient.
- Reaction bounds
-
User-defined constraints on the minimum and maximum allowable flux values for a particular metabolic reaction in a constraint-based model.
- Metabolite essentiality analysis
-
A metabolite-centric approach to determine essential components for cellular growth. To computationally test the essentiality of a metabolite, the consuming reactions of the particular metabolites are constrained to zero.
- Coupling constraints
-
Constraints that enforce strict relationships between model biochemical transformations, thereby connecting the fluxes for different cellular processes (such as transcription, translation, and tRNA and protein use for a metabolic reaction).
- Linear programming
-
A mathematical optimization technique that calculates the maximum or minimum value of a particular variable (that is, the objective function) on the basis of a set of linear constraints; an example of this is flux-balance analysis.
- Consensus sequences
-
Conserved sequences of nucleotides or amino acids that represent the target for a biomolecular event, often for proteins binding to the genome.
Rights and permissions
About this article
Cite this article
Bordbar, A., Monk, J., King, Z. et al. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet 15, 107–120 (2014). https://doi.org/10.1038/nrg3643
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3643
This article is cited by
-
Assumptions on decision making and environment can yield multiple steady states in microbial community models
BMC Bioinformatics (2023)
-
The effective deficiency of biochemical networks
Scientific Reports (2023)
-
Optimization of nutrient utilization efficiency and productivity for algal cultures under light and dark cycles using genome-scale model process control
npj Systems Biology and Applications (2023)
-
Genome-scale metabolic modeling of Aspergillus fumigatus strains reveals growth dependencies on the lung microbiome
Nature Communications (2023)
-
Identification of gene function based on models capturing natural variability of Arabidopsis thaliana lipid metabolism
Nature Communications (2023)