Abstract
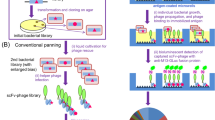
Here we describe a new method applying phage-displayed antibody libraries to the selection of antibodies against a single identified cell on a glass slide. This is the only described method that has successfully achieved selection of antibodies against a single rare cell in a heterogeneous population of cells. The phage library is incubated with the slide containing the identified rare cell of interest; incubation is followed by UV irradiation while protecting the target cell with a minute disc. The UV light inactivates all phages outside the shielded area by cross-linking the DNA constituting their genomes. The expected yield is between one and ten phage particles from a single cell selection. The encoded antibodies are subsequently produced monoclonally and tested for specificity. This method can be applied within a week to carry out ten or more individual cell selections. Including subsequent testing of antibody specificity, a specific antibody can be identified within 2 months.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Sorensen, M.D., Agerholm, I.E., Christensen, B., Kolvraa, S. & Kristensen, P. Microselection—affinity selecting antibodies against a single rare cell in a heterogeneous population. J. Cell Mol. Med. 14, 1953–1961 (2010).
Smith, G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 (1985).
McCafferty, J., Griffiths, A.D., Winter, G. & Chiswell, D.J. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 348, 552–554 (1990).
Bass, S., Greene, R. & Wells, J.A. Hormone phage: an enrichment method for variant proteins with altered binding properties. Proteins 8, 309–314 (1990).
Kristensen, P. & Winter, G. Proteolytic selection for protein folding using filamentous bacteriophages. Fold Des. 3, 321–328 (1998).
Orum, H. et al. Efficient method for constructing comprehensive murine Fab antibody libraries displayed on phage. Nucleic Acids Res 21, 4491–4498 (1993).
Rondot, S., Koch, J., Breitling, F. & Dubel, S. A helper phage to improve single-chain antibody presentation in phage display. Nat. Biotechnol. 19, 75–78 (2001).
Baek, H., Suk, K.H., Kim, Y.H. & Cha, S. An improved helper phage system for efficient isolation of specific antibody molecules in phage display. Nucleic Acids Research 30 (2002).
Hoogenboom, H.R. Selecting and screening recombinant antibody libraries. Nat. Biotechnol. 23, 1105–1116 (2005).
Pittenger, M.F. et al. Multilineage potential of adult human mesenchymal stem cells. Science 284, 143–147 (1999).
Zvaifler, N.J. et al. Mesenchymal precursor cells in the blood of normal individuals. Arthritis Res. 2, 477–488 (2000).
Asahara, T. et al. Isolation of putative progenitor endothelial cells for angiogenesis. Science 275, 964–967 (1997).
Nagrath, S. et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450, 1235–1239 (2007).
Fehm, T. et al. Cytogenetic evidence that circulating epithelial cells in patients with carcinoma are malignant. Clin. Cancer Res. 8, 2073–2084 (2002).
Johnson, K.L., Stroh, H., Khosrotehrani, K. & Bianchi, D.W. Spot counting to locate fetal cells in maternal blood and tissue: a comparison of manual and automated microscopy. Microsc. Res. Tech. 70, 585–588 (2007).
Kilpatrick, M.W. et al. Automated detection of rare fetal cells in maternal blood: eliminating the false-positive XY signals in XX pregnancies. Am. J. Obstet. Gynecol. 190, 1571–1578 discussion 1578–1581 (2004).
Marks, J.D. et al. Human antibody fragments specific for human blood group antigens from a phage display library. Biotechnology (NY) 11, 1145–1149 (1993).
Emmert-Buck, M.R. et al. Laser capture microdissection. Science 274, 998–1001 (1996).
Lu, H., Jin, D. & Kapila, Y.L. Application of laser capture microdissection to phage display peptide library screening. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 98, 692–697 (2004).
Ruan, W., Sassoon, A., An, F., Simko, J.P. & Liu, B. Identification of clinically significant tumor antigens by selecting phage antibody library on tumor cells in situ using laser capture microdissection. Mol. Cell Proteomics 5, 2364–2373 (2006).
Herzenberg, L.A. & Sweet, R.G. Fluorescence-activated cell sorting. Sci. Am. 234, 108–117 (1976).
de Kruif, J., Terstappen, L., Boel, E. & Logtenberg, T. Rapid selection of cell subpopulation-specific human monoclonal antibodies from a synthetic phage antibody library. Proc. Natl. Acad. Sci. USA 92, 3938–3942 (1995).
Fong, S.M., Lee, M.K., Adusumilli, P.S. & Kelly, K.J. Fluorescence-expressing viruses allow rapid identification and separation of rare tumor cells in spiked samples of human whole blood. Surgery 146, 498–505 (2009).
Letchford, J. et al. Isolation of C15: a novel antibody generated by phage display against mesenchymal stem cell-enriched fractions of adult human marrow. J. Immunol. Methods 308, 124–137 (2006).
Kurosaki, Y. et al. Pyrimidine dimer formation and oxidative damage in M13 bacteriophage inactivation by ultraviolet C irradiation. Photochem. Photobiol. 78, 349–354 (2003).
Celis, J.E. (ed.) Cell Biology: A Laboratory Handbook Third edn., (Elsevier Academic Press,, 2006).
Bussow, K. et al. A method for global protein expression and antibody screening on high-density filters of an arrayed cDNA library. Nucleic Acids Res. 26, 5007–5008 (1998).
Holt, L.J., Bussow, K., Walter, G. & Tomlinson, I.M. By-passing selection: direct screening for antibody-antigen interactions using protein arrays. Nucleic Acids Res. 28, E72 (2000).
Jakobsen, C.G., Rasmussen, N., Laenkholm, A.V. & Ditzel, H.J. Phage display-derived human monoclonal antibodies isolated by binding to the surface of live primary breast cancer cells recognize GRP78. Cancer Res. 67, 9507–9517 (2007).
Lee, C.M., Iorno, N., Sierro, F. & Christ, D. Selection of human antibody fragments by phage display. Nat. Protoc. 2, 3001–3008 (2007).
Christensen, B. et al. Studies on the isolation and identification of fetal nucleated red blood cells in the circulation of pregnant women before and after chorion villus sampling. Fetal Diagn. Ther. 18, 376–384 (2003).
Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Moutel, S. et al. A multi-Fc-species system for recombinant antibody production. BMC Biotechnol. 9, 14 (2009).
Jensen, K.B. et al. Functional improvement of antibody fragments using a novel phage coat protein III fusion system. Biochem. Biophys. Res. Commun. 298, 566–573 (2002).
Acknowledgements
We thank The Danish Council for Independent Research—Technology and Production Sciences (09-065063) and the FP6 EU project, PROTEOMAGE (LSHM-CT-2005-518230), for financial support. FCMB is thanked for partial financing of M.D.S.
Author information
Authors and Affiliations
Contributions
M.D.S. designed and conducted the experiments in the development of the method and wrote the manuscript. P.K. designed experiments, supervised the project and contributed to the writing of the manuscript. Both authors discussed the results and implications and commented on the manuscript at all stages.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Sørensen, M., Kristensen, P. Selection of antibodies against a single rare cell present in a heterogeneous population using phage display. Nat Protoc 6, 509–522 (2011). https://doi.org/10.1038/nprot.2011.311
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.311
This article is cited by
-
Development of a Phage Display Panning Strategy Utilizing Crude Antigens: Isolation of MERS-CoV Nucleoprotein human antibodies
Scientific Reports (2019)
-
Upregulation of Mrps18a in breast cancer identified by selecting phage antibody libraries on breast tissue sections
BMC Cancer (2017)
-
Using phage display selected antibodies to dissect microbiomes for complete de novo genome sequencing of low abundance microbes
BMC Microbiology (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.