Abstract
Bacterial artificial chromosomes (BACs) are widely used in studies of vertebrate gene regulation and function because they often closely recapitulate the expression patterns of endogenous genes. Here we report a step-by-step protocol for efficient BAC transgenesis in zebrafish using the medaka Tol2 transposon. Using recombineering in Escherichia coli, we introduce the iTol2 cassette in the BAC plasmid backbone, which contains the inverted minimal cis-sequences required for Tol2 transposition, and a reporter gene to replace a target locus in the BAC. Microinjection of the Tol2-BAC and a codon-optimized transposase mRNA into fertilized eggs results in clean integrations in the genome and transmission to the germline at a rate of ∼15%. A single person can prepare a dozen constructs within 3 weeks, and obtain transgenic fish within approximately 3–4 months. Our protocol drastically reduces the labor involved in BAC transgenesis and will greatly facilitate biological and biomedical studies in model vertebrates.
Similar content being viewed by others
Introduction
BAC transgenesis has been used extensively in a broad range of applications in mice, from studies of gene regulation to creating animal models of human disease1,2,3. Because BACs can hold genomic fragments as large as 300 kb, they often include the complete structure of a gene, including long-range cis-regulatory elements required for correct cell type–specific and temporal expression. Compared with small plasmid-based transgenes, BACs are generally more resistant to positional effects, presumably because of their larger size. BAC transgenic mice have uniquely advanced the analysis of distant cis-regulatory elements, rescue of mutant phenotypes and study of human disease-related genes1,2. BAC reporter transgenes have been created systematically by recombineering fluorescent reporters in bacteria to visualize specific tissues or cells in living mice4,5.
Given the tractability of zebrafish (Danio rerio) for vertebrate genetics, improving the frequency and reliability of BAC transgenesis in this model organism could have a number of important advantages. BAC transgenic zebrafish have been reported for more than 10 years6,7,8; however, systematic generation of BAC reporter lines has not been straightforward. BAC transgenic lines expressing live reporters such as GFP can enhance the usefulness of zebrafish in developmental and evolutionary studies, in modeling human disease9, targeted manipulation of neural circuits8 and drug screening9. As fertilization is external, live embryos are accessible to visualization and manipulation of specific cell types. Furthermore, genetic manipulations by targeted expression of apoptotic genes, neurotoxins and light-activated ion channels have become feasible through the Gal4/UAS10,11, LexA/Op12, Cre/lox13 or TetON14 conditional expression systems in zebrafish.
Traditionally, BAC transgenesis has been carried out by microinjection of naked DNA (purified DNA without associated proteins) in the fertilized zebrafish egg6 or mouse oocyte1. BAC integration in the genome occurs randomly via nonhomologous DNA end joining1. A number of reports suggest that BAC DNA microinjection into the cytoplasm of fertilized zebrafish eggs results in approximately 1–3% (or less) germline transmission6,7,8. Although there are no detailed reports of copy number and fidelity of BAC integration in transgenic zebrafish, extensive studies of BAC transgenesis in mice have shown that approximately half of BAC integrations result in either a single-copy BAC insertion or insertion of multicopy BAC concatemers at a single genomic locus; the remaining carry between 5 and 48 copies of the BAC in various orientations15. Concatemeric transgenes are generally associated with silencing, instability and genetic lesions both inside and around the transgenes16,17, potentially limiting important experimental applications. The conditional deletion of a particular DNA sequence within a BAC using Cre-loxP–mediated recombination2 is one such application. In particular, when tandem loxP sites are placed within a BAC flanking a gene or cassette and the BAC transgene is a concatemer, unwanted deletions may occur upon Cre-mediated recombination. Therefore, more reliable methods for BAC transgenesis in zebrafish have been desired.
In the past two decades, a number of transgenesis methods have been developed for zebrafish, yet most are not applicable to BAC transgenesis because of restrictions in DNA cargo capacity (Table 1). Microinjection of naked DNA, in which a linearized construct is introduced into the cytoplasm of one-cell-stage embryos18,19,20,21, is the simplest and easiest method, but suffers from low integration rates and is unreliable as discussed above. Inclusion of an 18-bp I-SceI meganuclease recognition site in the plasmid DNA and co-injection with I-SceI protein was found to substantially enhance integration rates for small (∼5 kb) constructs22; whether this is useful for enhancing integration of BAC constructs in zebrafish is not known. Retroviral vectors have also been successfully used for transgenesis in zebrafish, particularly for genome-wide insertional mutagenesis23,24,25,26; however, retroviral vectors have a very limited cargo capacity (<8 kb)27 and their application in the laboratory is labor intensive. More recently, attention has turned to transposable elements, including mariner28, Tol2 (ref. 29) and Sleeping beauty30. Among them, Tol2 appears to have the highest rate of genomic integration in the germ lineage (Table 1) and is now widely used for transgenesis and forward genetics, including insertional mutagenesis31,32. Furthermore, we recently demonstrated that Tol2 has a surprisingly large cargo capacity (more than 50 kb) and can carry efficiently BAC inserts into the zebrafish and mouse genomes33.
The medaka Tol2 element is a DNA-type transposon that is active in a wide variety of vertebrates31,32,33,34,35. When cis-sequences from the left and right ends of Tol2 are placed on either side of a DNA insert, the insert can be reliably integrated into the genome via a transposase (TP)-dependent cut-and-paste mechanism34,36. By inverting these minimal Tol2 ends, 150 bp on the left and 200 bp on the right, and placing them ∼1 kb apart on a large BAC plasmid, we have found that any insert can be efficiently excised by the Tol2 TP in fertilized zebrafish eggs or mouse oocytes33. By using this inverted minimal Tol2 cassette (iTol2 cassette), we have observed a very high success rate in the integration of BAC transgenes into the zebrafish genome33,37, regardless of insert size (range 35–230 kb) or source (five BAC libraries tested from two species). Stable germline BAC integrations are observed in ∼15% of the injected founder fish (range 5–20%). Notably, reporter expression in the BAC transgenic fish matches the expression pattern of the endogenous gene.
Here we report a step-by-step protocol, including new iTol2 cassettes (iTol2-galactokinase (galK), iTol2-ampicillin (amp) and iTol2-kanamycin (kan)), reagents and recent examples from our work that illustrate in detail how to generate BAC transgenic zebrafish efficiently using recombineering technology38 and the Tol2 transposon system. Our protocol complements conventional naked DNA injection by providing increased efficiency and higher reliability to deliver single-copy BAC integrations. This protocol should greatly expand the use of BAC transgenesis in zebrafish, aiding clean and intact BAC integrations and more reliable tools for analysis of gene regulation, comparative genomics and targeted gene expression.
Overview of the procedure
Generation of BAC transgenic zebrafish with the Tol2 system consists of three stages. A flowchart in Figure 1 outlines these stages and the experimental procedures involved. In the first stage (Steps 1–33), a Tol2-compatible BAC plasmid is constructed in E. coli by recombineering. A BAC clone covering the genomic region of interest is retrieved by searching online databases. The clone is grown in a special strain of E. coli in order to introduce by homologous recombination a reporter gene (such as GFP) at the locus of interest and the iTol2 cassette into the plasmid backbone. In the second stage (Steps 34–48), the BAC transgene is microinjected into fertilized eggs together with Tol2 TP RNA. In the third stage (Steps 49–60), the injected fish are raised to sexual maturity and screened for transmission of the BAC transgene to the germline by PCR genotyping or fluorescence sorting. Below we describe the background and implementation of the procedure.
Obtaining BAC clones. In general, as the regulatory elements of any gene may be scattered over long distances and located both upstream and downstream of the gene39, it is wise to start with the largest possible genomic clones available (at least 100 kb). If the gene is too large to be contained in a single BAC, ideally several overlapping BAC clones should be obtained. These clones can be modified and tested in parallel to identify the most appropriate BAC clone for stable transgenesis. Clones are available from at least eight zebrafish genomic libraries, including CHORI-211, CHORI-73, CHORI-1073, DanioKey and DanioKey Pilot (Table 2). Fosmid clones in the CHORI-1073 library can also be modified by recombineering because the inserts are in the single-copy vector pCCFOS1.
BAC recombineering in E. coli. To modify a BAC clone, DNA sequences in the BAC are exchanged by bacteriophage-mediated homologous recombination systems in E. coli38,40,41,42,43,44. BAC plasmid DNA is introduced by electroporation into the bacterial strain SW102 (derived from DY380), which harbors a defective λ-Red prophage on the genome that contains the heat-inducible recombinase functions, and a precise deletion of the galK gene (ΔgalK)45. This strain allows both single-step antibiotic selection and two-step positive/negative galK selection45. A linearized PCR product (the cassette) containing a donor sequence and 50 bp homologies to the target sequence on each end is introduced by electroporation into BAC-containing cells41,45. The λ-Red–encoded Gam function prevents degradation of the PCR product, whereas Exo or Redα and Beta or Redβ mediate recombination. Because Redα/Redβ are under the tight control of the temperature-sensitive λ repressor (allele cI857)40,41, recombination can be induced by shifting the cultures to 42 °C for 15 min, leading to precise exchange of the target sequence45,46,47,48,49.
iTol2 cassette. To facilitate integration of the BAC construct into the genome, a cassette containing the minimal sequences required for Tol2 transposition must be placed inside the BAC plasmid. As 200 bp on the left (L200) and 150 bp on the right end (R150) of the Tol2 transposon are sufficient for efficient transposition in vivo36 (Fig. 2a), we created the iTol2 cassette, which consists of the inverted L200 and R150 sequences flanking the galK-, amp- or kan-resistance genes. To prepare linear double-stranded iTol2 cassette DNA for recombineering, piTol2-galK, piTol2-amp and piTol2-kan (Fig. 2a) are used as templates for PCR.
(a) Schematic of the medaka Tol2 transposon and design of the iTol2 cassettes. Red triangles represent the left (L200) and right (R150) minimal ends of Tol2 required for transposition. Gray boxes and overlying lines in Tol2 represent the exons of TP mRNA. In piTol2-amp, piTol2-kan and piTol2-galK, the L and R ends of Tol2 have been inverted flanking the amp-resistance (ampR), kan-resistance (kanR) and galK genes, respectively. The prokaryotic em7 promoter is included in piTol2-galK. Plasmids carry the Spec-resistance gene (SpecR). (b) Schematic of vectors used for PCR amplification of galK and kanamycin recombineering. Kanamycin selection plasmids encoding GFP, Gal4FF, Gal4VP16 or Cre contain the SV40 pA (gray box), where the kan is flanked by FRT sites.
Reporter gene cassette. Typically, a cassette containing a reporter gene (such as GFP) is placed in a coding exon within the BAC to monitor the expression of a gene of interest. We have constructed several plasmids containing a number of reporter gene cassettes that serve as templates for PCR (Fig. 2b). In these plasmids, the reporter gene (such as GFP) is fused to a strong transcription termination signal (i.e., simian virus 40 pA) and an antibiotic-resistance gene (kan) flanked by FRT sites. Two of these plasmids encode the Gal4FF10 and Gal4-VP16 (refs. 50,51) transcriptional activators, and another the Cre recombinase52. Gal4 drives transcription of any target gene located downstream of its target UAS sequence50,51, and Cre will delete any sequence located between two adjacent loxP sites52. To restrict the activity of Gal4 or Cre in space and time, the cassettes are typically placed under the control of tissue-specific promoters in the BAC4,5.
Introducing the iTol2 cassette into the BAC clone. The iTol2 cassette with 50-bp homologies on each end is amplified by PCR and inserted at a fixed location in the BAC plasmid—either a loxP or lox511 site, which is present in the plasmid backbone of most BAC libraries (Fig. 3a). After transformation of the PCR product into BAC-containing cells, and induction of recombineering functions, colonies containing the BAC plasmid with the iTol2-amp cassette (Tol2-BAC plasmid) will grow on ampicillin medium overnight (Fig. 3a).
(a) iTol2 cassette and kanamycin selection cassette recombineering. The iTol2-amp linear fragment with 50-bp overhangs (small arrows represent primers) matching a sequence flanking a loxP or lox511 site is introduced into the BAC clone, which contains a gene of interest (gene X). After selection on ampicillin plates, bacteria containing the resulting Tol2-BAC plasmid are electroporated with a PCR product containing GFP and kanamycin flanked by FRT sites, with 50-bp homologies to the first and second exons in the gene. After selection on ampicillin and kanamycin plates, colonies containing the Tol2-BAC reporter are readily obtained. (b) Introducing a reporter gene into the Tol2-BAC by galK recombineering. A PCR-amplified galK product with 50-bp homologies to the first and second exons in the gene is electroporated into cells containing the Tol2-BAC reporter. After selection on galactose-only medium, Gal+ colonies containing the Tol2-BAC:galK are recovered. Next, these colonies are electroporated with a linear PCR product encoding a reporter gene (GFP) to replace the galK cassette. After negative selection on DOG medium, colonies containing the Tol2-BAC:GFP reporter are recovered.
Introducing the reporter gene into the BAC clone. The reporter gene is introduced into the Tol2-BAC plasmid using antibiotic selection (Fig. 3a) or two-step galK selection (Fig. 3b). A kan reporter gene cassette (such as GFP-pA-FRT-kan-FRT) is amplified by PCR, together with 50-bp homologies to the first exon and the first intron (or second exon) of the target gene. After transformation of Tol2-BAC-containing cells with the PCR product and induction of recombineering functions, kan+ recombinants (Tol2-BAC:FRT-kan-FRT) are recovered on Luria broth (LB)-ampicillin/kanamycin plates the next day (Fig. 3a). Although it is not essential to do so, the kan gene can be excised later on from the Tol2-BAC plasmid by induction of FLPase in E. coli38,45. For galK selection, a 1.2 kb galK gene, including the prokaryotic em7 promoter (Fig. 2b) and 50-bp homologies to the BAC target sequence, is amplified by PCR and electroporated into Tol2-BAC-containing cells45. Only cells containing galK (gal+) will grow on galactose minimal medium after 3 d (Fig. 3b). To replace galK with a reporter gene, a PCR product containing the gene of interest (such as GFP) and 50-bp homologies is transformed into gal+ cells. After induction of recombineering functions, cells are plated on minimal medium containing 2-deoxy-D-galactose (DOG), a galactose analog that, after phosphorylation by galK, becomes toxic45. Only bacteria containing the Tol2-BAC reporter plasmid without galK (Gal–) will survive on DOG-containing medium after 3 d (Fig. 3b).
Microinjection of the Tol2 -BAC plasmid and stable transgenesis. To integrate the BAC construct in the genome, the Tol2-BAC plasmid and synthetic Tol2 TP mRNA are co-injected into the cytoplasm of one-cell-stage embryos53 (Fig. 4). Efficient integration of the Tol2-BAC construct in the genome is strongly dependent on the activity of TP33. To increase the translation efficiency of TP mRNA, we recently generated a synthetic TP gene with codon usage optimized for zebrafish (zTP, Fig. 4a). Injection of zTP mRNA leads to reliable germline transmission of Tol2-BAC transgenes in ∼20% of injected fish. To confirm that TP is functioning properly and that the BAC plasmid DNA is intact, a simple PCR excision assay is performed on 10-h-old injected embryos (Fig. 4b). TP excises the BAC DNA insert upon binding to the iTol2 sequences, creating an 8-bp duplication at the target site34,36 (Fig. 4c). If excision occurs, the bacterial selection gene inside the iTol2 cassette is released, and then circularized presumably by self-ligation33,34 (Fig. 4b,c). Ideally, nine of ten embryos show an excision product, and if so all injected embryos are raised to sexual maturity. However, if the BAC transgene encodes a fluorescent reporter, it is recommended to raise separately those showing the strongest and most even fluorescence signals from the rest, as stronger signals may be correlated with higher integration rates (M.L.S., unpublished observations). To identify germline carriers among injected (founder) fish, the progeny are screened by PCR or fluorescent sorting. We recommend screening at least 50 (but preferably 100) embryos from each founder.
(a) A synthetic zebrafish codon-optimized Tol2 TP cDNA (zTP) was cloned into the pCS2 expression vector. After linearizing this vector with NotI, zTP mRNA is synthesized in vitro from the SP6 promoter. (b) The Tol2-BAC and TP mRNA are co-injected into the cytoplasm of the one-cell embryo. A subset of the injected embryos are grown for 10 h, digested with proteinase K and processed for a PCR excision assay. The PCR products amplified from the excised iTol2 cassette (see below) are examined on a 1% (wt/vol) agarose gel. A strong 336-bp band (arrowhead) is detected in embryos co-injected with BAC and TP RNA (+TP mRNA) but not without TP (−TP mRNA). M, 1 kb DNA ladder. (c) Schematic of Tol2-dependent excision and integration of the Tol2-BAC transgene in the genome. TP protein (black circles) binds to the iTol2 ends in the Tol2-BAC plasmid (pTol2-BAC), leading to excision of the amp gene (blue arrow) that is circularized by ligation. This excision product can be amplified by PCR (using the forward (f) and reverse (r) primers). The Tol2 ends of the excised BAC plasmid align with a random target site (asterisk) in the genomic DNA, followed by stable integration and duplication of the 8-bp target sequence (asterisks). AmpR, amp resistance.
Advantages, applications and limitations
Two major advantages of Tol2-mediated BAC transgenesis over existing methods are increased efficiency of germline transmission and greater reliability of obtaining single-copy integrations in the genome (Table 1). BAC transgenic fish expressing fluorescent proteins in specific cells and tissues have a wide range of unique applications in basic and applied fields of biology and medicine. These include live imaging of cells and subcellular structures in complex tissues and organs such as the nervous system8. Other applications include rescue of mutant phenotypes, mechanistic studies of human disease9, targeted cell ablation and manipulation of neural circuits54, drug screening9 and functional analysis of gene regulatory elements6,37. Because the iTol2 sequences can be added in a single recombineering step to many BACs at once, our protocol can uniquely facilitate the systematic generation of BAC transgenic fish. One limitation of our protocol is that clones maintained in multicopy plasmids such as cosmids (30–50 kb) are not readily modifiable by recombineering38. In addition, target regions containing repetitive or incorrect sequences can pose obstacles to recombineering. Although we are not aware of any size limitation on Tol2-mediated BAC transgenesis, this has yet to be determined.
Experimental design
Choice of BAC clone. Given the number of vertebrates with sequenced genomes, there are in principle a large number of possible sources of BAC clones for transgenesis in zebrafish. Although most mammalian promoters are not likely to work in transgenic zebrafish55, BACs from closely related teleost fish, such as the pufferfish Fugu rubripes, are known to work properly in zebrafish37 and are useful tools for comparative genomic studies. Such BAC clones may be useful when zebrafish BACs are not available for a particular genomic region or when this region contains excessive repeats. Table 3 lists information regarding BAC libraries from several model fish species with sequenced genomes.
Choice of recombineering system. Two recombineering systems commonly used are the λ-Red system (the one used in this protocol) and the RecET system from the Rac prophage38. Both systems permit recombination using double-stranded fragments with short (40–60 bp) homologies40,41,42,43,44. To supply the recombination functions, there are two options. One is to use a bacterial strain that harbors the genes encoding these functions (such as SW102). Another option is to supply a plasmid that contains these genes (such as pSIM18 (ref. 56)). The latter could be useful to avoid the preparation of BAC plasmid DNA before recombineering. On the other hand, new bacterial strains containing the recombineering functions, such as SW105 and SW106 (derivates of SW102), offer advantages as well, such as the presence of arabinose-inducible Cre or FLP in the E. coli genome, which permit the excision of cassettes flanked by loxP or FRT sites, respectively45.
Cassette design and recombineering procedure. Construction of Tol2-BAC transgenes may require several rounds of recombineering and cassettes. Thus, the size of the cassette, selection marker (amp, kan and galK), target sequence and procedure should be carefully considered. Although homologous exchange of DNA up to 2 kb is straightforward, larger cassettes (>5 kb) require cloning longer homology arms around 200–500 bp because of the lower efficiency of homologous recombination38. Subcloning is more laborious and large cassettes are more prone to mutagenesis during PCR. If a cassette has a generic function (such as the iTol2 cassette), it should be inserted at a fixed location in the BAC plasmid, thus simplifying the process and saving costs. As long as the target site does not contain repetitive sequences, it should be possible to insert the cassette almost anywhere in the BAC.
Strain for Tol2 -BAC microinjection. In principle, the BAC construct should be microinjected into fertilized eggs derived from the healthiest zebrafish strain (with the highest survival rate) such as the wild-type strains TAB or TL. However, alternative strains may be used to facilitate screening of transgenic carriers or to avoid delays due to breeding in the future. If the aim is to create a BAC transgenic line expressing Gal4, the Tol2-BAC construct can be injected directly into the homozygous UASGFP strain10 to select fluorescent-positive-injected embryos/larvae (carrying both BAC:Gal4 and UASGFP). Another useful strain is nacre, a mitfa-recessive mutant lacking melanophore pigmentation57, which is transparent and facilitates live imaging of internal tissues.
Materials
REAGENTS
-
Zebrafish (Danio rerio): wild-type strains (TAB or TL) and transgenic Tg(UAS:GFP) fish10
Caution
All animal experiments should adhere to relevant institutional ethics guidelines.
-
Aquatic facility with 2- and 12-liter tanks (Aqua Schwarz or equivalent companies)
-
Anesthetic (ethyl 3-aminobenzoate methanesulfonate salt (Sigma, cat. no. A5040)
-
SW102 bacteria: SW102 (mcrA Δ(mrr-hsdRMS-mcrBC) ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7649 galU rspL nupG Φ80dlacZΔM15 λcI857 (cro-bioA)◊tetΔgalK) harbors a defective λ-prophage and deletion of the ΔgalK45 (obtained from NCI-Frederick) (http://web.ncifcrf.gov/research/brb/recombineeringInformation.aspx)
-
BAC clones purchased from BACPAC Resouces Center or ImaGenes (Table 2)
-
galK plasmid: pBluescript SK-pEm7-galK ampicillin resistance (AmpR)45 (available from NCI-Frederick)
-
iTol2 cassette plasmids: pCR8GW-iTol2-amp, pCR8GW-iTol2-kan and pCR8GW-iTol2-galK available from M.L.S. or K.K. on request (see Supplementary Data for plasmid map and full sequence)
-
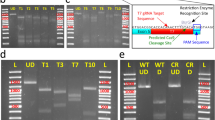
Kan cassette plasmids: pBSK-GFP-pA-FRT-kan-FRT, pCR8GW-Gal4FF-pA-FRT-kan-FRT, pCR8GW-Gal4-VP16-pA-FRT-kan-FRT, pCR8GW-Cre-pA-FRT-kan-FRT available from M.L.S. on request (see Supplementary Data for plasmid map and full sequence)
-
Tol2 TP plasmid: zebrafish codon-optimized Tol2 TP, pCS2-zT2TP available from K.K. on request (see Supplementary Data for plasmid map and full sequence)
-
Primers for amplifying iTol2 cassettes (Table 4; Choose primer according to the source of the BAC clone (Table 2). Order 50 nmol with PAGE purification (Sigma) and resuspend in 10 mM Tris-Cl, pH 8.5, at 100 μM.)
Table 4 Primers and adapter sequences. -
Gene-specific primers for recombineering reporter gene cassettes and primers for confirming homologous recombination (REAGENT SETUP and Table 4; order 50 nmol with PAGE purification for oligos longer than 50 bp and 25 nmol for up to 50 bp. Resuspend in 10 mM Tris-Cl, pH 8.5, at 100 μM.)
-
Primers for sequencing reporter genes after recombineering (Table 4; order 25 nmol (Sigma), desalted and resuspend in 10 mM Tris-Cl, pH 8.5, at 100 μM)
-
Oligos for adapter-ligation (ADL) PCR (Table 4; order 25 nmol (Sigma), desalted and resuspend in 10 mM Tris-Cl, pH 8.5, at 100 μM)
-
Primers for single-embryo PCR excision assay (Table 4; order 25 nmol (Sigma), desalted and resuspend in 10 mM Tris-Cl, pH 8.5, at 100 μM)
-
Plasmid miniprep kit (Qiagen, cat. no. 27106)
-
QIAquick gel extraction kit (Qiagen, cat. no. 28706)
-
Nucleobond BAC 100 kit (Macherey-Nagel, cat. no. 740579)
-
Ethanol (99.7%)
-
Isopropanol (Sigma, cat. no. I9516)
Caution
It is very flammable and irritating to eyes. Inhalation may cause dizziness and nausea.
-
Phenol-chloroform-isoamyl alcohol mixture 25:24:1 (Sigma, cat. no. 77617)
Caution
It is toxic by inhalation, ingestion or contact with skin.
-
Chloroform (Sigma, cat. no. C0549)
Caution
It is carcinogenic. Wear gloves.
-
Ammonium chloride (Sigma, cat. no. A9434)
Caution
It is harmful if swallowed and can be irritating to eyes.
-
Ammonium sulfate (Sigma, cat. no. A4418)
-
Calcium chloride (Sigma, cat. no. C1016)
-
Methylene blue hydrate (Sigma, cat. no. 28514)
Caution
It is harmful if swallowed and can be irritating to the eyes, respiratory system and skin.
-
Lithium chloride (Sigma, cat. no. 62478)
-
Magnesium sulfate heptahydrate (Sigma, cat. no. M1880)
-
Potassium chloride (Sigma, cat. nos. P9333 and RNase-free P9541)
-
Potassium phosphate, monobasic (Sigma, cat. no. P5655)
-
Sodium chloride (Sigma, cat. no. S7653)
-
Sodium phosphate, dibasic (Sigma, cat. no. S7907)
-
Ferrous sulfate heptahydrate (Sigma, cat. no. F8048)
Caution
It is harmful if swallowed.
-
Potassium hydroxide (Sigma, cat. no. P5958)
Caution
It is toxic and highly corrosive.
-
D-biotin (Sigma, cat. no. 47868)
-
Glycerol, min 99% (Sigma, cat. no. G6279)
-
D-(+)-galactose (Sigma, cat. no. G0750)
-
2-Deoxy-D-galactose (Sigma, cat. no. 31050)
-
L-Leucine (Sigma, cat. no. L8912)
-
Ampicillin sodium salt, cell culture tested (Sigma, cat. no. A0166)
Caution
Inhalation and skin contact may cause allergic reactions.
-
Chloramphenicol (Sigma, cat. no. C-0378)
Caution
It is carcinogenic.
-
Kanamycin sulfate (Invitrogen, cat. no. 11815)
-
Tetracycline hydrochloride (Sigma, cat. no. T3383)
Caution
It is irritating to the eyes, respiratory system and skin.
-
Spectinomycin dihydrochloride pentahydrate (Sigma, cat. no. 85555)
-
Minimal salts, M9
-
Sterile H2O purified, deionized and 0.22 μm filtered (Milli-Q, Millipore)
-
Bacto-tryptone (Difco, cat. no. 0123-01-1)
-
Bacto-yeast extract (Difco, cat. no. 0127-05-3)
-
Bacto-agar (Difco, cat. no. 0140-01)
-
MacConkey agar base (Difco, cat. no. 281810)
-
Agarose (Sigma, cat. no. A5093)
-
Tris base (Sigma, cat. no. T1503)
-
Glacial acetic acid (Sigma, cat. no. 320099)
Caution
It is flammable and highly corrosive.
-
Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA; Sigma, cat. no. E5134)
-
Ethidium bromide solution, 10 mg ml−1 (Sigma, cat. no. E1510)
Caution
It is toxic by inhalation and it causes damage to DNA.
-
BlueJuice gel loading buffer (10×; Invitrogen, cat. no. 10816-015)
-
DNA ladder (1 Kb; Invitrogen, cat. no. 15615-016)
-
λ DNA/HindIII fragments (Invitrogen, cat. no. 15612-013)
-
mMESSAGE mMACHINE SP6 kit (Ambion, cat. no. 1340)
-
Mini Quick Spin RNA columns (Roche, cat. no. 11814427001)
-
RNA Ladder (0.5–10 Kb; Invitrogen, cat. no. 15623-200)
-
NorthernMax kit for running denaturing RNA gel (Invitrogen, cat. no. AM1940)
Caution
Loading dye, gel and buffer contain formaldehyde. It is toxic by inhalation, ingestion and skin contact. It is carcinogenic.
-
Nuclease-free water (Invitrogen, cat. no. AM9930)
-
Phenol red sodium salt (Sigma, cat. no. 114537)
Caution
It is irritating to the eyes, respiratory system and skin. Wear gloves when handling it.
-
Proteinase K (Sigma, cat. no. P2308)
Caution
It is irritating to the eyes, respiratory system and skin.
-
Sodium dodecyl sulfate (Invitrogen, cat. no. 15525-017)
-
Sodium acetate buffer solution, pH 5.2 (3M; Sigma, cat. no. S7899)
-
Expand high-fidelity PCR system or a similar kit with proofreading ability (Roche, cat. no. 11681834001)
-
GoTaq Flexi DNA polymerase for quick PCR screening (Promega, cat. no. M8305)
-
PCR nucleotide mix (dNTPs), 10 mM each (Roche, cat. no. 11581295001)
-
Restriction enzymes (AluI, DpnI, MboI, BglII, NotI, SalI, SpeI, XbaI and XhoI) and T4 DNA ligase (Takara and NEB)
-
DNA sequencing, BigDye terminator v3.1 cycle sequencing kit; 5× sequencing buffer for diluting enzyme; HiDi formamide (Applied Biosystems)
EQUIPMENT
-
Thermal cycler and accessories for PCR (MyCycler, Bio-Rad)
-
DNA electrophoresis system, submerged horizontal (Sub-Cell Systems, Bio-Rad)
-
Spectrophotometer for determining DNA concentration (NanoDrop, Thermo Scientific)
-
Spectrophotometer for measuring bacterial optical density at 600 nm (OD600) (DU640, Beckman)
-
Benchtop refrigerated centrifuge, capacity 0.5–50 ml (Eppendorf, cat. no. 5805000017, model 5804R, Eppendorf)
-
Benchtop centrifuge (Eppendorf, cat. no. 5424000410, model 5418)
-
Constant temperature incubators set at 32 °C and 37 °C (Medinor, cat. no. 15110)
-
Constant temperature dry heat blocks set at 42–95 °C (Medinor, cat. no. D1105)
-
Shaking water bath, 210 r.p.m, with temperature control (Grant)
-
Sterile Petri dishes, 92 × 16 mm (Sarstedt, cat. no. 82.1472.001)
-
Sterile Petri dishes for microinjection, 60 × 15 mm (Falcon, cat. no. 351007)
-
Sterile tubes for PCR (0.2 ml; VWR, cat. no. 732-0547)
-
Eight-strip PCR tube (0.2 ml) and flat 8-cap strips for PCR (Bio-Rad, cat. nos. TLS-0801, TCS-0803)
-
PCR tube with flat cap (0.5 ml; VWR, cat. no. 732-0675)
-
Sterilized Eppendorf tubes, 3810 (1.5 ml; Eppendorf, cat. no. 700-5239)
-
Sterile tubes for storage of bacterial stocks (2 ml; Sarstedt, cat. no. 72.694.406)
-
SafeSeal micro tubes (2 ml; Sarstedt, cat. no. 72.695)
-
Bacterial round-bottom tubes with cap (14 ml; BD Biosciences, cat. no. 352059)
-
Falcon tubes (15 ml; VWR, cat. no. 734-0451)
-
Falcon tubes (50 ml; VWR, cat. no. 734-0448)
-
Duran Erlenmeyer narrow-neck flasks (50 ml; Sigma, cat. no. Z232785)
-
Gilson Pipetman set and filtered tips (VWR, cat. nos. 732-2225, 732-2236,732-2207)
-
Hand-held electric Pipetman for 1–10 ml volume (Medinor P2000)
-
Electroporation machine, MicroPulser (Bio-Rad, cat. no. 165-2100)
-
GenePulser electroporation cuvettes, 0.1-cm gap (Bio-Rad, cat. no. 1652089)
-
Plastic mating boxes, 24.5 × 15 × 16.5 cm (Spawn Box 3, Aqua Schwarz)
-
Glass capillaries for microinjection (GC-1, Narishige)
-
KIMAX-51 capillaries, 0.8–1.10 × 100 mm (Sigma, cat. no. KIM-34502-99-20EA)
-
Microloader tips for loading glass capillaries (Eppendorf, cat. no. 5242 956.003)
-
Micropipette capillary puller (Sutter Instruments, cat. no. P-87)
-
Dow Corning Sylgard 184 elastomer kit (VWR, cat. no. 634I65S)
-
Dumont No. 5 forceps, 0.05 mm × 0.01 mm (Fine Science Tools, cat. no. 11254-20)
-
Pressure microinjection system with pipette holder (Applied Scientific Instrumentation, MPPI-3)
-
Dissecting stereomicroscope (SMZ645, Nikon)
-
Fluorescent stereomicroscope equipped with GFP/RFP filters (MZFLIII, Leica)
-
Computer with web browser and other software for analysis of genomic DNA sequences and primer design (DNA Strider, ApE Plasmid Editor v2.0.30 and Amplify 3×)
-
Capillary electrophoresis instrument (e.g., Applied Biosystems 3130xl or similar)
REAGENT SETUP
Zebrafish
-
Adult fish are maintained at a density of 10–30 fish per 12-liter tank with a 14-h light:10-h dark cycle at 28.5 °C.
Caution
All work must comply with relevant ethical guidelines on animal experiments.
Chloramphenicol
-
Chloramphenicol is prepared by dissolving 12.5 mg ml−1 in 99.7% (vol/vol) ethanol. Store at −20 °C in 1-ml aliquots for up to 1 year.
Caution
Wear gloves and a dust mask when handling powder and solution.
Kanamycin
-
Kanamycin is prepared by dissolving 25 mg ml−1 in H2O. Store at −20 °C in 1-ml aliquots for up to 1 year.
Ampicillin
-
Ampicillin is prepared by dissolving 50 mg ml−1 in H2O. Store at −20 °C in 1-ml aliquots for up to 1 year.
Tetracycline
-
Tetracycline is prepared by dissolving 12.5 mg ml−1 in 50% (vol/vol) ethanol. Store at −20 °C in 1-ml aliquots for up to 1 year. Protect from light.
Spectinomycin
-
Spectinomycin is prepared by dissolving 50 mg ml−1 in H2O. Store at −20 °C in 1-ml aliquots for up to 1 year.
LB medium
-
Mix 10 g of Bacto-tryptone, 5 g Bacto-yeast and 5 g of NaCl in 1 liter of distilled H2O. Autoclave and store at 25 °C for up to 6 months.
LB-agar plates
-
Sterile Petri dishes (92 mm × 16 mm) containing 1% (wt/vol) Bacto-agar with one or several antibiotics: chloramphenicol, 12.5 μg ml−1; kanamycin, 25 μg ml−1; ampicillin, 50 μg ml−1; and tetracycline, 12.5 μg ml−1. Store at 4 °C for up to 2 months.
SW102 electrocompetent cells
-
Streak bacteria onto an LB-tetracycline plate to obtain single colonies. Pick a single colony and grow overnight in 5 ml of LB-tetracycline at 32 °C, with shaking at 210 r.p.m. Prepare a glycerol stock by combining 500 μl of this culture and 500 μl of 30% (wt/vol) glycerol in a 2-ml sterile tube; store at −80 °C for up to 1 year. Starting from this stock, prepare electrocompetent cells as described in Box 1.
M9 medium
-
Mix 6 g of Na2HPO4, 3 g of KH2PO4, 1 g of NH4Cl and 0.5 g of NaCl in 1 liter of sterile H2O. Autoclave and store at 25 °C in a glass bottle for up to 1 year.
M63 medium (5×)
-
Mix 10 g of (NH4)2SO4, 68 g of KH2PO4 and 2.5 mg of FeSO4·7H2O in 1 liter of sterile H2O; adjust pH to 7 with KOH. Autoclave and store at 25 °C in a glass bottle for up to 1 year.
M63 minimal medium plates
-
Dissolve 15 g Bacto-agar in 800 ml of distilled H2O, sterilize and then add 200 ml of 5× M63 medium, 1 ml of 1 M MgSO4·7H2O, 10 ml of 20% (wt/vol) galactose, 5 ml of 0.2 mg ml−1 biotin (0.2 μm sterile filtered), 4.5 ml of 10 mg ml−1 leucine (heated and 0.2-μm sterile filtered) and antibiotic. Pour into ∼40 92-mm Petri dishes and store at 4 °C for up to 2 months.
DOG minimal medium plates
-
Dissolve 15 g Bacto-agar in 800 ml of distilled H2O, sterilize, and then add 200 ml of 5× M63 medium, 1 ml of 1 M MgSO4.7H2O, 10 ml of 20% (wt/vol) glycerol, 10 ml of 20% (wt/vol) DOG, 5 ml of 0.2 mg ml−1 biotin, 4.5 ml of 10 mg ml−1 leucine and antibiotic. Pour into ∼40 92-mm Petri dishes and store at 4 °C for up to 2 months.
MacConkey agar plates
-
Dissolve 5 g of agar in 500 ml of distilled H2O, 5 ml of 20% (wt/vol) galactose, sterilize, cool and add antibiotic. Pour into ∼30 92-mm Petri dishes and store at 4 °C for up to 2 months.
E3 medium
-
Mix 20 ml of 5 M NaCl, 3.4 ml of 1 M KCl, 6.6 ml of 1 M CaCal2, 6.6 ml of 1 M MgSO4 and 2 ml of 0.05% (wt/vol; optional) methylene blue in 20 liters of sterile H2O. Adjust pH to 7. Store at 25 °C for up to 2 months.
TE buffer
-
Combine 10 mM Tris, pH 8.0, and 1 mM EDTA. Store at 25 °C for up to 6 months.
Tricaine solution
-
Dissolve 20 mg of Tricaine in 100 ml of E3 medium. Freshly prepare on the day of use.
Phenol red solution
-
Dissolve 2.5% of (wt/vol) phenol red in nuclease-free water. Store at −20 °C in 1-ml aliquots for up to 1 year.
Embryo lysis buffer
-
Mix 10 mM Tris, 10 mM EDTA and 200 μg ml−1 proteinase K. Freshly prepare on the day of use.
Genomic DNA extraction buffer
-
Combine 10 mM Tris-HCl, pH 8.2; 10 mM EDTA; 200 mM NaCl; 0.5% (wt/vol) SDS; and 200 μg ml−1 proteinase K. Freshly prepare on the day of use.
TAE (10×)
-
Mix 48.4 g of Tris base, 11.42 ml of glacial acetic acid and 3.72 g of EDTA in 1 liter of H2O. Store at 25 °C for up to 6 months.
Gene-specific primers
-
For recombineering, design 82–90-mer single-stranded PCR oligos to amplify galK, GFP, Gal4FF, Gal4VP16 or Cre and the kan recombineering cassettes (Fig. 2b) using the sequences listed in Table 4. Also design 25–30-mer oligos to confirm the recombination events by PCR, by choosing sequences that are approximately 50–100 bp away from the BAC target site. To replace an open reading frame in the BAC, one cassette primer should contain sequence annealing to at least 50 bp surrounding and upstream of the expected translation start, whereas the second primer should contain sequence annealing to an exon/intron at least 500 bp downstream from the first primer. In general, primers should have a melting temperature of >55 °C and a minimum of a 50-bp homology to the BAC sequence. Free online software such as Amplify 3× (http://engels.genetics.wisc.edu/amplify/) can be used for designing the primers.
ADL-PCR adapters
-
Mix the following in 1.5-ml Eppendorf tubes: 50 μl of 100 μM ASS(GATC) or ASS(CTAG), 2 μl of T4 polynucleotide kinase, 10 μl of 10× buffer, 10 μl of 10 mM ATP and 28 μl of sterile H2O. Incubate at 37 °C for 1 h and at 75 °C for 5 min, add 50 μl of 100 μM ALS, incubate at 94 °C for 1 min and at 60 °C for 10 min, cool down to 25 °C, precipitate with 99.7% (vol/vol) ethanol, spin down for 10 min at 16,000g, wash with 70% (vol/vol) ethanol and resuspend in a final volume of 50 μl of 10 mM Tris-Cl, pH 8.5. Store at −20 °C for up to 6 months.
EQUIPMENT SETUP
Embryo microinjection dish
-
Fill the lid of a Petri dish (60 × 15 mm) with enough Sylgard (prepared according to the manufacturer's instructions) just to cover four glass capillaries (Kimax-51, ∼3.5 cm long, outer diameter 1 mm) placed at equal distance and parallel to each other. Alternatively, prepare a 1% (wt/vol) agarose ramp with a glass plate as described previously53.
Microinjection capillaries
-
Pull glass capillaries (Narishige GC-1) with a Micropipette puller (e.g., Sutter P-87, heat = 645, pull = 163, vel = 145 and time = 50) to obtain a long and fine needle; break with fine forceps to create a ∼2–3-μm tip. Prepare immediately before microinjection.
Microinjection setup
-
Set up the ASI MPPI-3 pressure injection system with foot pedal external trigger according to the manufacturer's instructions. A source of compressed air with at least 20 psi is required.
Procedure
Identification of BAC clones
Timing 1 d
-
1
Open the Ensembl Zv9 (http://www.ensembl.org/Danio_rerio/Info/Index) or ZFIN (http://zfin.org/cgi-perl/gbrowse/current/) websites in your web browser and type the gene name or perform a search using BLAST options.
-
2
In Ensembl, open the 'Gene' page of your gene of interest and click on 'Location' to open 'chromosome view'. Select 'Configure this page' from the left panel. This will open a new window inside the page. In this window, click 'Other DNA alignments' under the Main panel. Enable 'BAC ends' and 'FOS ends' to see genomic clones in your region of interest.
-
3
Locate 'BAC ends' that appear as blue squares within the expanded genomic region. Click on those BACs (unfilled rectangles flanked by the blue squares) whose ends cover the gene of interest to obtain the Ensembl prefix (e.g., zC175D15.za). It is preferable to use a BAC clone in which the gene of interest is located in the middle of the BAC, as it is more likely that such a clone will contain most, if not all, of the cis-regulatory elements required for the full and correct expression pattern of that gene.
-
4
Align the entire coding sequence of the gene with the contig, scaffold or BAC clone sequence to identify unambiguously all exons, introns and the first methionine. In some cases, you can use the NCBI Genome Project browser to identify clones or 'trace' sequences that may not have been annotated yet in Ensembl. We recommend using computerized DNA analysis programs such as DNA Strider to carry out sequence alignments.
-
5
Order the BAC, PAC or fosmid (FOS) clones. To order the clones, you must find the original library clone name and number corresponding to the Ensembl prefix obtained in Step 3 above (see also Table 2). For example, a clone with the name zC175D15 in Ensembl belongs to the CHORI-211 library (prefix zC), clone number 175D15. If your gene is larger than 100 kb and there are several BAC or FOS clones that cover the region of interest, order two clones that overlap.
Critical Step
Pay careful attention to the correct identification of the gene and BAC clones, particularly if your genomic region has not been well annotated.
Critical Step
If possible, avoid excessively large BACs (>200 kb), as these are more prone to breakage during BAC DNA preparation and microinjection steps. In addition, keep in mind that the presence of unusually high GC-rich sequences or many repeats might lead to toxicity that could compromise recombineering or transgenesis steps.
Preparation of BAC-containing competent cells
Timing 4 d
-
6
Streak a small amount of bacteria from the BAC agar stab shipped by the supplier to a fresh LB-chloramphenicol agar plate to obtain single colonies.
-
7
Pick two individual colonies using a plastic filtered tip on a P20 pipette and transfer them into 14-ml round-bottom tubes containing 5 ml of LB-chloramphenicol. Grow overnight at 32 °C with shaking at 210 r.p.m.
-
8
The next day, prepare BAC DNA from the bacterial cultures using a quick miniprep protocol (Box 2) and ensure that DNA is resuspended in 50 μl of sterile water. Remember to retain 500 μl of the culture to prepare a glycerol stock by mixing 500 μl of culture and 500 μl of sterile 30% (wt/vol) glycerol; store at −80 °C indefinitely.
-
9
Digest BAC plasmid DNA to check its size and integrity as follows:
Table 6
Component
Amount (μl)
Final
BAC plasmid DNA
25
Unknown
Takara (10×) H or M buffer
4
1×
SalI (20 U μl−1) or SpeI (10 U μl−1)
1
Variable
Sterile water
To 40
-
10
Add 4 μl of gel loading dye (10× BlueJuice) to the reaction and run the sample in a 0.8% (wt/vol) agarose gel. Run the gel for ∼1 h at 90 V. Photograph the gel under UV light (400 nm–315 nm) and observe the digestion pattern. Compare the observed pattern with the one expected from the available genomic DNA sequence using the free web tool at http://tools.neb.com/NEBcutter2/.
Critical Step
If unexpected band sizes are observed upon restriction digestion, we recommend repeating DNA isolation and rechecking several more colonies before proceeding.
-
11
Thaw SW102 electrocompetent cells stored as 25-μl aliquots at −80 °C. Place cells on ice (always keep close to 0 °C).
-
12
Transfer 25 μl of SW102 cells to a prechilled 1.5-ml tube. Add 1–5 μl (approximately 200–500 ng) of BAC DNA to the cells, mix gently by tapping and incubate for 5–10 min on ice.
-
13
Transfer the cells to a prechilled 0.1-cm gap cuvette and keep it on ice. Quickly transfer the cuvette to the electroporation holder and apply 1.8 kV, 25 μF and 200 Ω resistance, with a time constant of 5 msec, by pressing the pulse button under the Ec1 setting in the BioRad MicroPulser electroporator.
Caution
Ensure that the outside of the cuvette is dry before applying a current.
Critical Step
Ensure that bacteria are quickly transferred from ice to the electroporation holder. Cells must be cold during electroporation.
-
14
Immediately add 1 ml of LB to the cuvette, pipette cells up and down and transfer to a 14-ml round-bottom tube. Incubate at 32 °C for 1 h with shaking at 210 r.p.m.
-
15
Transfer cells to a 1.5-ml Eppendorf tube. Spin at 14,000g for 30 s. Remove most of the supernatant, leaving 100 μl behind. Resuspend by tapping a few times (do not vortex). Spread the transformed bacteria evenly on LB-chloramphenicol agar plates (as the BAC clone carries the CmR gene). Incubate the plates overnight at 32 °C.
Recombineering the iTol2 cassette into the BAC plasmid
Timing 2 d
-
16
Dilute the piTol2-amp, piTol2-kan or piTol2-galK plasmids (Fig. 2a) to 1 ng μl−1 and prepare aliquots of the iTol2 cassette primers at 50 μM in H2O. For CHORI-211 and CHORI-73 BAC libraries, use primers ptarbac_itol2_fw and ptarbac_itol2_rev; for CHORI-1073, DanioKey and DanioKey Pilot, use primers pindigobac_itol2_fw and pindigobac_itol2_rev (Table 2 and for further information).
-
17
Combine the following reagents for a 100-μl PCR in a 0.2-ml sterile tube:
Table 7
Component
Amount (μl)
Final
piTol2-amp plasmid DNA (1 ng μl−1)
1
1 ng
Expand high-fidelity buffer (10×) with MgCl2
10
1×
10 mM dNTP mixture (10 mM)
2
0.3 mM
Primer itol2_fw (50 μM)
2
1 μM
Primer itol2_rev (50 μM)
2
1 μM
Expand high-fidelity enzyme (3.5 U μl−1)
1
3.5 U
Nuclease-free water
To 100
-
18
Run the PCR with the following conditions optimized for Roche PCR enzymes on a Bio-Rad machine (MyCycler):
Table 8
Cycle number
Denature
Anneal
Extend
1
94 °C, 2 min
2–30
94 °C, 30 s
58 °C, 30 s
72 °C, 2 min
31
72 °C, 5 min
-
19
Remove the template plasmid DNA by adding it to 2 μl of DpnI to the PCR product and incubating it at 37 °C for 6–8 h.
Critical Step
It is essential to ensure complete digestion of the plasmid DNA template by DpnI. Otherwise, many false-positive colonies will arise in Step 32.
-
20
Purify the PCR product by adding 10 μl of 10× BlueJuice, split the sample and load 50 μl each into two wells in a 1% (wt/vol) agarose TAE gel. Perform gel extraction using the QIAquick gel extraction kit according to the manufacturer's instructions. Use 10 μl of a 100 ng μl−1 1-kb ladder to check the size of PCR products, which should be 1416 bp for iTol2-amp, 1368 bp for iTol2-kan and 1,662 bp for iTol2-galK. Collect PCR product with sterile H2O into a sterile 1.5-ml tube at a final concentration of 0.2–1 μg μl−1. Typically, this PCR product is enough for recombineering 20–30 BACs.
Pause point
PCR product can be stored at −20 °C for several weeks.
-
21
Pick two single SW102 colonies containing the BAC (from Step 15) into 14-ml round-bottom tubes containing 5 ml of LB-chloramphenicol and shake overnight at 32 °C, 210 r.p.m.
-
22
Transfer 500 μl of overnight SW102:BAC culture into 25 ml LB-chloramphenicol (prewarmed to RT) in a 50-ml flask. Grow cells at 32 °C, 210 r.p.m., until OD600 reaches 0.55–0.6. This takes approximately 3–4 h.
Critical Step
Do not harvest cells under or overgrown before proceeding to the next step. It is crucial that the OD600 is within 0.55–0.6. If not, restart Step 22.
-
23
Prepare ice/sterile water slush in a Styrofoam box, and place inside it for each flask: 50 ml of autoclaved Milli-Q water, 2 15-ml Falcon tubes, 2 1.5-ml Eppendorf tubes. Place 6 10-ml pipettes and 2 electroporation cuvettes in a refrigerator at 4 °C.
-
24
Heat-shock the SW102 cells with BAC at 42 °C for exactly 15 min, shaking at 210 r.p.m. (or by hand every 4 min).
Critical Step
The cells must not be heat shocked longer than 15 min and should be shaken periodically to ensure even heat transfer.
-
25
Place the cells in a flask (from Step 24) on ice/water slush immediately. Allow to cool for 5 min. Mix gently by inverting up/down and transfer 10 ml from each to a 15-ml Falcon tube. Store the rest at 4 °C.
-
26
Centrifuge the tubes for 5 min at 4,500g in an Eppendorf centrifuge precooled to 0 °C. Immediately after the spin is completed,transfer the tubes to ice/water slush.
-
27
Carefully but rapidly drain out the supernatant by inverting the tubes without disturbing the pellet. Resuspend pellet by adding 10 ml of prechilled autoclaved Milli-Q water using a chilled 10-ml pipette.
Critical Step
Carefully resuspend by keeping tubes inside the ice/water slush at this and all steps, because warming or harsh treatment of cells will reduce the success of recombineering drastically.
-
28
Centrifuge for 5 min at 4,500g in an Eppendorf centrifuge precooled to 0 °C. Immediately after the spin is completed, transfer the tubes to ice/water slush. Repeat Steps 27 and 28.
-
29
Carefully but rapidly drain out the supernatant by inverting the tubes without disturbing the pellet. This time, ensure that virtually all supernatant is drained out, except for ∼50 μl left at the bottom, by inverting the tube on tissue paper and tapping it a few times. Transfer this 50 μl of competent cells to a new prechilled 1.5-ml tube and keep it on ice until you are ready for electroporation.
Critical Step
The cells should be used immediately, but within 30 min at the latest.
-
30
Mix 50 μl of cells and 1–2 μg of iTol2 cassette and electroporate as described in Steps 12 and 13.
-
31
Recover cells with 1 ml of LB and grow at 32 °C for 1 h. Keep a 100-μl aliquot of these cells for plating (Step 32). Centrifuge the remaining 900 μl, then remove all supernatant except the 'bottom' 100 μl and resuspend the pellet by tapping a few times.
-
32
Plate 100 μl of the cells before and after centrifugation on LB-chloramphenicol-ampicillin agar plates for iTol2-amp and on LB-chloramphenicol-kanamycin for iTol2-kan. To avoid false positives, make replica plates from the same colonies on both LB-chloramphenicol-kanamycin and LB-chloramphenicol-spectinomycin agar plates. For iTol2-galK, plate transformed cells on M63 minimal plates with chloramphenicol. Incubate overnight at 32 °C for iTol2-amp and iTol2-kan and 3 days for iTol2-galK.
Recombineering a reporter gene into the Tol2-BAC plasmid
-
33
To introduce a reporter gene into the BAC, we recommend one of the following two options, depending on the aim of the experiment. Option A consists of recombineering a kanamycin cassette and overnight selection on LB-agar medium. This option is ideal for modifying many BACs in parallel. Option B involves recombineering a galK cassette, 3 d of positive selection on minimal medium, followed by a second round of recombineering with a reporter gene and 3 d of negative selection on DOG minimal medium. This option is best for making a translational fusion and for creating precise deletions or point mutations inside a BAC.
-
A
Recombineering a reporter gene by kanamycin selection
-
i
Amplify the GFP-pA-FRT-kan-FRT, Gal4FF-pA-FRT-kan-FRT, Gal4VP16-pA-FRT-kan-FRT or Cre-pA-FRT-kan-FRT cassette (Fig. 2b and Supplementary Data) according to the conditions used in Steps 17 and 18, except that the extension time should be increased to 2 min 40 s. Use 1 ng of the plasmid DNA and gene-specific primers (geneX_GFP_fw, geneX_Gal4FF_fw, geneX_Gal4-VP16_fw or geneX_Cre_fw and geneX_frt-kan-rev) designed to match 50 bp upstream and downstream of the chosen target site on the BAC plasmid (see REAGENT SETUP and Table 4).
-
ii
Remove the plasmid DNA template by digesting the PCR product with 2 μl of DpnI. Incubate it at 37 °C for 6–8 h.
-
iii
Precipitate 95 μl of PCR product by adding 5 μl of 5 M LiCl and 300 μl of 99.7% (vol/vol) ethanol. Mix well and place for 30 min at −20 °C. Run the remaining 5 μl of PCR product on a 1% (wt/vol) TAE-agarose gel to check the success of PCR.
Pause point
PCR product can be stored at −20 °C overnight.
-
iv
Centrifuge the precipitated PCR product at 19,300g for 15 min at 4 °C.
-
v
Carefully wash the pellet once with 1 ml of 70% (vol/vol) ethanol.
-
vi
Dry the pellet at 25 °C for 5–10 min.
-
vii
Resuspend in 10 μl of nuclease-free water.
-
viii
Use 1 μl of the suspension to measure DNA concentration with a spectrophotometer. Concentration is normally in the range approximately 2–5 μg μl−1.
-
ix
Pick a colony using a plastic filtered tip on a P20 pipette from SW102 cells harboring Tol2-BAC (from Step 32) and transfer it into a 14-ml round-bottom tube containing 5 ml of LB-chloramphenicol-ampicillin. Grow overnight at 32 °C with shaking at 210 r.p.m.
-
x
Add 500 μl of the overnight culture into 25 ml of LB-chloramphenicol-ampicillin. Grow cells at 32 °C to OD600 = 0.55–0.6 (3–4 h) with shaking at 210 r.p.m.
-
xi
Prepare competent cells as described in Steps 23–29.
-
xii
Transform the cells with kan cassette DNA (5–10 μg) by electroporation as described in Steps 12 and 13.
-
xiii
Recover cells and plate on LB-chloramphenicol-ampicillin-kanamycin agar plates according to Steps 14 and 15. To avoid false positives, make replica plate colonies on LB-chloramphenicol-ampicillin-spectinomycin agar plates.
-
xiv
Pick 10–12 colonies from the LB-chloramphenicol-ampicillin-kanamycin agar plates (which did not grow on spectinomycin) and prepare plasmid DNA according to Box 2.
-
xv
Analyze DNA by restriction enzyme digestion with SalI or SpeI. Run it on a 0.8% (wt/vol) agarose gel and photograph the gel under UV illumination. Compare the digestion pattern of the original BAC, Tol2-BAC and final Tol2-BAC reporter plasmids (Fig. 5).
Figure 5: Restriction analysis of BAC plasmid DNA before and after recombineering the iTol2-amp and Gal4-VP16 cassettes. BAC DNA miniprep was prepared as described in Box 1. A volume of 20 μl from each prep was digested with SalI and separated on a 1% (wt/vol) agarose gel. BAC, Tol2-BAC and Tol2-BAC:Gal4 DNA from BAC clones CHORI-211-266O5, CHORI-1073-398L10, CHORI-73-313L2 are shown. In some cases, new bands or changes in band size may be detected from one recombineering step to the next (arrows). λ, lambda DNA ladder; M, 1-kb DNA ladder.
-
xvi
Confirm homologous recombination by PCR amplification using gene-specific primers (see REAGENT SETUP) and sequence the reporter gene using GFP_seqF, Gal4FF_seqF, Gal4-VP16_seqF or Cre_seqF and Kan_seqR primers, depending on the cassette (Table 4).
Timing 2 d
-
i
-
B
Recombineering a reporter gene by galK selection
-
i
Amplify the galK cassette (Fig. 2b) by PCR using 1 ng of the pgalK plasmid and gene-specific primers (geneX_galK_fw and geneX_galK_rev) with homologies designed to match 50 bp upstream and downstream of the chosen target site on the BAC plasmid (see REAGENT SETUP and Table 4). Set up PCR reactions and conditions as described in Steps 17 and 18.
-
ii
Digest the PCR product to remove the plasmid DNA template and purify as described in Steps 19 and 20.
Pause point
PCR product can be stored at −20 °C for several weeks.
-
iii
Pick a colony using a plastic filtered tip on a P20 pipette from SW102 cells harboring Tol2-BAC (Step 32) and transfer it into a 14-ml round-bottom tube containing 5 ml of LB with chloramphenicol and ampicillin. Grow overnight at 32 °C with shaking at 210 r.p.m.
-
iv
Add 500 μl of the overnight culture into 25 ml of LB with chloramphenicol and the iTol2 cassette antibiotic (ampicillin or kanamycin). Grow cells at 32 °C to OD600 = 0.55–0.6 (∼4 h) with shaking at 210 r.p.m.
-
v
Prepare competent cells as described in Steps 23–29.
-
vi
Transform the cells with galK cassette DNA (25-50 ng) by electroporation as described in Steps 12 and 13.
-
vii
Transfer the bacteria to a 14-ml bacterial round-bottom tube with 1 ml of LB; incubate at 32 °C for 1 h with shaking at 210 r.p.m.
-
viii
Wash the bacteria twice with 1× M9 medium. Resuspend the pellet in 1 ml of 1× M9 medium, and plate 10, 100 and 250 μl of cells onto M63 minimal medium plates with chloramphenicol (and ampicillin or kanamycin). Incubate at 32 °C for 3 d.
-
ix
Pick colonies and streak on MacConkey agar plates with chloramphenicol. Pick single bright red (Gal+) colored colonies and grow them in 5 ml of LB-chloramphenicol-ampicillin or LB-chloramphenicol-kanamycin at 32 °C overnight.
-
x
Amplify the reporter gene (e.g., GFP, Gal4FF, Gal4-VP16 or Cre) by PCR using the plasmids in Figure 2b as templates and gene-specific primers (e.g., geneX_GFP_fw and geneX_pA_rev in Table 4) containing the same 50-bp homologies used for the galK primers in Step 33B(i). Digest the PCR product with DpnI and purify by gel extraction as described in Steps 19 and 20. Alternatively, use double-stranded oligos containing a point mutation, deletion or insertion to create the appropriate modification at the target site38.
-
xi
Introduce 1–2 μg of PCR product or double-stranded oligos into Gal+ cells from Step 33B(ix) according to the electroporation procedure described in Steps 12 and 13.
-
xii
Transfer the cells to 10 ml of LB and incubate at 32 °C for 4.5 h with shaking at 210 r.p.m.
-
xiii
Wash the bacteria twice with 1× M9 medium. Resuspend the pellet in 1 ml of 1× M9 medium, and plate 10, 100 and 250 μl of cells onto DOG minimal medium plates with chloramphenicol and ampicillin or kanamycin. Incubate at 32 °C for 3–4 d.
-
xiv
Prepare plasmid DNA from 10 to 12 colonies according to Box 2 and analyze BAC DNA as described in Steps 33A(xvi) and 33A(xvii).
Timing 6 d
-
i
-
A
Microinjection of the Tol2-BAC DNA and TP RNA
Timing 3 d
-
34
Prepare recombineered BAC DNA using the Nucleobond BAC 100 kit according to the manufacturer's instructions. Next, purify DNA by phenol-chloroform-isoamyl alcohol extraction, precipitate with one-tenth volume of 3 M sodium acetate (pH 5.2) and 3 volumes of 99.7% (vol/vol) ethanol, rinse once with 70% (vol/vol) ethanol and resuspend in nuclease-free water at 250 ng μl−1. To resuspend the BAC DNA, flick the tube gently several times and let it rest at 25 °C for at least 1 h. Keep a small aliquot of the BAC DNA at 4 °C for immediate use and store the rest at −20 °C. Avoid repeated freeze/thawing of the BAC DNA before microinjection.
Caution
Phenol is toxic by inhalation, ingestion or contact with skin. Wear suitable gloves and work in a fume hood.
Critical Step
Never vortex or pipette BAC DNA up and down, as this will lead to shearing.
Critical Step
Avoid transferring any of the bottom phenol layer and any material at the interface during DNA extraction. Impurities and/or phenol are highly toxic to embryos and will likely result in mortality upon microinjection (Step 44).
-
35
Digest pCS2-zT2TP (Fig. 3a) with NotI and purify by phenol-chloroform-isoamyl extraction followed by chloroform extraction. Ethanol precipitate, rinse once with 70% (vol/vol) ethanol and dissolve in nuclease-free water at the concentration of ∼1 μg μl−1.
-
36
Use mMESSAGE mMACHINE SP6 kit to synthesize Tol2 TP mRNA from 1 μg of the linearized template according to the manufacturer's instruction.
-
37
Add nuclease-free water to the synthesized mRNA to a final volume of 100 μl and purify the sample with a mini quick-spin RNA column according to the manufacturer's instruction. Adjust the volume of the eluted solution to 135 μl. Add 15 μl of ammonium acetate stop solution and extract the mRNA once with an equal volume of phenol-chloroform-isoamyl alcohol and then with an equal volume of chloroform. Transfer the aqueous phase to a new tube.
-
38
Add an equal volume of isopropanol, mix well, chill the mixture at least for 15 min at −20 °C and centrifuge the mixture at 4 °C for 15 min at maximum speed. Carefully remove the supernatant and resuspend the precipitated mRNA in 50 μl of nuclease-free water. Approximately 30 μg of mRNA will be synthesized. Take an aliquot and adjust the concentration to 250 ng μl−1 for microinjection. Store it in 5-μl aliquots at −80 °C.
Caution
Isopropanol is very flammable and irritating to eyes; inhalation may cause dizziness and nausea.
Pause point
Transposase mRNA can be stored at −80 °C for several months.
-
39
Ensure that MPPI-3 microinjection system is working properly, including the air supply (Fig. 6), and prepare a dish for holding the embryos in place during microinjection (see EQUIPMENT SETUP and Fig. 6b).
Figure 6: Microinjection setup and breeding and raising of transgenic zebrafish. (a) Nikon SMZ645 stereomicroscope with backlight illumination, embryo dish, forceps and MPPI-3 pressure injection system is shown. The arrow points to the foot pedal trigger. (b) The Sylgard dish for embryo injection and the needle holder. Trenches (1 mm wide) are indicated by arrows in which embryos are lined up (white dashed line) and covered with E3 medium. Scale bar, 5 mm. (c) Injection needle filled with BAC/RNA phenol red solution (asterisk) in the MPPI-3 holder. Scale bar, 2.5 cm. (d) Mating box with males (top) and females (bottom) before mating. A green nylon ball is placed inside the box as a spawning stimulus. Scale bar, 2.4 cm. (e,f) A 2-liter tank system for raising young larvae (e) and a 12-liter tank system for raising adult fish (f).
-
40
Place one or two male and one or two female zebrafish in separate compartments of a 3-liter mating box in the late afternoon after feeding (Fig. 6d). Prepare enough mating boxes to set up four to six pair matings to obtain enough eggs for several rounds of injection in a given morning.
-
41
The next morning, prepare the BAC DNA/RNA injection mixture in a 0.5-ml RNAse-free tube as follows, mix by gently flicking the tube several times and always keep on ice:
Table 9
Component
Amount (μl)
Final
Tol2-BAC plasmid DNA (250 ng μl−1)
4
1 μg
Tol2 transposase RNA (250 ng μl−1)
4
1 μg
KCl (0.4 M)
10
0.2 M
Phenol red solution (2.5%, wt/vol)
2
0.25%
Critical Step
Do not vortex or pipette BAC DNA mixture up and down, as this will lead to breakage of the Tol2-BAC plasmid.
-
42
Prepare fine needles for microinjection (Fig. 6c) by pulling a glass capillary, and break the tip with fine forceps. Load 3 μl of the DNA/RNA mixture into the glass capillary needle using a P20 pipette and Microloader tips. Attach the capillary (Fig. 6c) to the needle holder of the MPPI-3 pressure injection system (Fig. 6a). Set the pressure to 20 psi and adjust the pulse duration to 50 ms.
-
43
Mate fish in one 3-liter mating box (from Step 40) at a time by placing male and female fish together in the upper compartment and wait until they lay eggs. Collect eggs into Petri dishes with E3 medium. Microinjection should be carried out at the one-cell stage, namely within 20–30 min after fertilization.
-
44
Line up ∼60 newly fertilized zebrafish eggs (obtained from paired matings in Step 43) inside an embryo injection dish (Fig. 6b) containing E3 medium. Once each embryo is in place, rapidly penetrate the chorion of the one-cell-stage embryo with the tip of the needle (Fig. 6c, asterisk), and then move slowly into the boundary between the yolk and cytoplasm. Once the needle is inside the cytoplasm, inject enough solution to fill at least one-tenth of the cytoplasmic volume. Repeat Step 43 and inject 100–200 embryos for each BAC construct. Save some uninjected embryos as controls to check the health of the clutch. Incubate the injected embryos in E3 medium at 28.5 °C in Petri dishes until the desired age for use in the following steps.
Critical Step
It is crucial to inject the DNA/RNA mix directly into the cell (not into the yolk) at the one-cell stage. The phenol red included in the solution helps monitor the location and amount of solution injected. If necessary, an eyepiece graticule and micrometer slide can be used to measure the relative volume of the injected solution. A useful site describing the use of the graticule can be found at http://www.cavehill.uwi.edu/FPAS/bcs/courses/Biology/BIOL2053/2053proj/biol2053estimating%20size.htm.
Confirmation of Tol2-mediated BAC excision in injected embryos
Timing 2 d
-
45
Transfer eight injected embryos from Step 44 at 10 h post fertilization (hpf) to wells in an eight-strip PCR tube. Remove as much E3 medium as possible. Add 50 μl of embryo lysis buffer. Incubate at 50 °C overnight. Inactivate the enzyme at 95 °C for 10 min.
Critical Step
It is crucial to inactivate the proteinase K at 95 °C for 10 min. Failure to do so will result in no PCR amplification in the following steps.
Pause point
Embryo extracts can be stored at −20 °C for several weeks.
-
46
Combine the reagents tabulated below to make an 8× master mix for the PCR 'excision assay'. Below we list the primers for the iTol2-amp cassette, but details of excision primers for the iTol2-kan and iTol2-galK cassettes are also provided in Table 4. Dispense 20 μl of this mix per tube and add 1 μl of the embryo extract from Step 45.
Table 10
Component
Amount (μl)
Final
Buffer (5×) with green loading dye
32
1×
MgCl2 (25 mM)
16
2.5 mM
dNTP mixture (10 mM)
3.2
0.2 mM
Primer iT2amp-exc168F (50 μM)
3.2
1 μM
Primer iT2amp-exc168R (50 μM)
3.2
1 μM
GoTaq Flexi DNA polymerase (5 U μl−1)
1.6
8 U
Nuclease-free water
To 160
-
47
Run the PCR with the following conditions:
Table 11
Cycle number
Denature
Anneal
Extend
1
94 °C, 2 min
2–35
94 °C, 30 s
55 °C, 30 s
72 °C, 1 min
36
72 °C, 5 min
-
48
Check 10 μl of the PCR samples by agarose gel electrophoresis (2% (wt/vol) agarose in TAE). The excision product (336 bp) should be detected from embryos injected with both the Tol2-BAC plasmid and the TP mRNA but not from embryos injected with Tol2-BAC plasmid only (Fig. 4b).
Critical Step
If no excision product is detected, the injected embryos should be discarded. Check the quality of BAC DNA and TP RNA and repeat injection (or restart from Step 34).
Selection and rearing of Tol2-BAC–injected fish
Timing 3–4 months
-
49
If injected embryos (F0) are expected to express fluorescent proteins such as GFP, screen the injected embryos/larvae for fluorescence under a dissection microscope equipped with UV light and appropriate filters. Record the tissues and cells that show fluorescence. Separate embryos/larvae with very bright and/or even fluorescence signals and grow them separately from the rest. Stronger fluorescence can be correlated with higher integration rates (M.L.S., unpublished observations). Transfer up to 30 injected larvae at 5 days post fertilization (dpf) from Step 44 into a 200-ml Pyrex glass beaker. Grow larvae in beakers until 12 dpf, and then transfer them to 2-liter tanks until they are 2 months old (Fig. 6d). Finally, raise them until they are sexually mature (3–4 months) in 12-liter tanks (Fig. 6e).
Screening of Tol2-BAC–injected fish
Timing 3 d
-
50
Identify F0 fish with integrations in the germline, by mating single fish as described below and screening their progeny either by visualization of the fluorescence signal (option A) or by PCR screening (option B).
-
A
Fluorescence screening
-
i
Place single male or female injected fish in mating boxes in the late afternoon, at least 1 h after feeding. If the BAC carries the GFP reporter, mate the injected fish with wild-type fish. If the BAC carries Gal4, mate the injected fish with UASGFP fish10.
-
ii
Collect eggs the next morning in E3 medium and examine them every day from 10 hpf to 5 dpf for fluorescence (Fig. 7).
Figure 7: Generation and identification of Tol2-BAC transgenic fish. (a) Schematic of the ∼70-kb Tol2BAC-Fugu evx1:Gal4FF plasmid containing the exon 2:Gal4FF fusion downstream of the hoxA cluster. Gray box is the plasmid backbone. (b,c) Transient mosaic expression Fugu evx1:Gal4FF after co-injection with TP RNA into homozygous UAS:GFP fertilized eggs at 10 h post fertilization (hpf) (b) and 4 days post fertilization (dpf) (c). GFP expression is observed widely at 10 hpf, but becomes very restricted to commissural neurons by 4 dpf (arrowheads). Transient 'ectopic' expression is observed in muscle cells. (d,e) Double-transgenic Tol2BAC-Fugu evx1:Gal4FF; UAS:GFP larvae obtained by mating two founder fish (nos. 18 (d) and 43 (e)) and homozygous UAS:GFP. A total of 5% (2 of 40) founder fish were germline carriers. GFP expression is localized to neurons in the hindbrain (hb) and spinal cord (sc). The arrowhead shows GFP expression in cloaca consistent with endogenous evx1 expression. (f,g) Tol2BAC-fgf24:GFP-1 (∼165 kb) (f) and Tol2BAC-fgf24:GFP-2 (∼140 kb) (g) contain a fusion of GFP and exon 1 of fgf24. (h,i) Transient mosaic GFP expression at 10 hpf (h) and 30 hpf (i). Arrowheads point to the otic vesicle. (j) Representative embryo from stable transgenic Tol2BAC-fgf24:GFP-2 at 24 hpf. Arrowheads point to the main sites of GFP expression that recapitulate endogenous fgf24 expression (see Supplementary Fig. 1). A total of 20% (5 of 25) and 22% (2 of 9) of fish injected with Tol2BAC-fgf24:GFP-1 and Tol2BAC-fgf24:GFP-2 were germline carriers. Scale bars, b, ∼1980 μm; c,e, ∼250 μm; h, ∼1875 μm; i, ∼278 μm; and j, ∼400 μm.
-
iii
Set aside founder fish whose progeny show stable expression of the reporter gene in the correct pattern33,37 (e.g., Fig. 7a,d,e; see Supplementary Fig. 1 for comparison of reporter gene and endogenous expression).
-
iv
Raise the fluorescence-positive F1 progeny to adulthood and maintain stable lines by outcrossing.
Critical Step
A minimum of 50 (and ideally 100) embryos should be observed from each mating. On average, there is a strong correlation between transmission frequency in the F1 progeny and transgene copy number. As Tol2 tends to generate single-copy BAC integrations, very few embryos are expected to show fluorescence in a clutch.
-
i
-
B
PCR screening
-
i
Place a single male or female injected fish in mating boxes in the late afternoon, at least 1 h after feeding.
-
ii
Collect 20–50 embryos in 200 μl of embryo lysis buffer and follow Steps 46–47 with primers specific for each reporter gene (e.g., GFP_seqF, Gal4FF_seqF, Gal4-VP16_seqF or Cre_seqF and pA_seqR in Table 4).
-
iii
Run a 1–2% (wt/vol) agarose gel and visualize PCR products under UV light.
-
iv
Set aside injected fish whose embryo clutches were PCR positive. Mate these fishes again as described in Step 50B(i) and grow their progeny until adulthood.
-
i
-
A
Analysis of Tol2-BAC integrations (optional)
Timing 3 d
-
51
Extract genomic DNA from transgenic adult zebrafish tail fins according to Box 3; include extracts from an adult wild-type control and an adult UASGFP control fish. Southern blot hybridization can be used to determine the actual number of transposon insertions as described previously33,53. However, as Tol2 typically generates one or two BAC integrations in the zebrafish germline, it is relatively easy to retrieve the Tol2-genomic junctions by inverse PCR or ADL-PCR. Steps 52–60 describe how to retrieve the genomic junctions by ADL-PCR and determine the BAC integration site. The full procedure is illustrated in Figure 8 along with typical results.
Figure 8: Analysis of Tol2-BAC integrations. (a) A section of the tail fin from adult Tol2BAC-Fugu evx1:Gal4FF fish was cut for DNA extraction. (b) Genomic DNA is digested with GATC or CTAG restriction enzymes (e.g., MboI) to create Tol2 genomic left (L) and right (R) fragments. After ligation of GATC or CTAG oligo 'adapters', the Tol2 genomic junctions are amplified in two steps (first and second PCR) with nested primers Ap1 and Ap2. The final products can be sequenced directly with Tol2-specific primers. (c,d) Schematic of the genomic integration sites of Tol2BAC-Fugu evx1:Gal4FF in two independent lines33. The duplicated sequence of the 8-bp target site is indicated. chr, chromosome; ntt4, orphan sodium- and chloride-dependent neurotransmitter transporter (solute carrier family 6 member 17); cng3, cyclic nucleotide gated channel beta 3.
-
52
Set up a digest as tabulated below, and then incubate at 37 °C for 1 h:
Table 12
Component
Amount (μl)
Final
Genomic DNA from Step 51
Variable
1 μg
NE (10×) buffer 4
2
1×
MboI (5 U μl−1)
1
10 U
Nuclease-free water
To 20
-
53
Inactivate the enzyme at 70 °C for 15 min, set up a ligation reaction as tabulated below and incubate it at 16 °C overnight. The next day, inactivate the ligase at 70 °C for 15 min.
Table 13
Component
Amount (μl)
Final
MboI-digested DNA from Step 52
2
Unknown
GATC adapter (see REAGENT SETUP)
2
Unknown
Takara (10×) ligase buffer
2
1×
T4 DNA ligase (350 U μl−1)
1
350 U
Nuclease-free water
To 20
Pause point
Inactivated ligation reactions can be stored at −20 °C overnight.
-
54
Dilute the entire ligation reaction tenfold by adding 180 μl of H2O. Set up the first PCR as tabulated below:
Table 14
Component
Amount (μl)
Final
Tenfold-diluted DNA from Step 53
2
Unknown
Expand high-fidelity buffer (10×) with MgCl2
5
1×
dNTP (10 mM) mixture
1.5
0.3 mM
Primer Ap1 (50 μM)
1
1 μM
Primer 175L-out or 150R-out (50 μM)
1
1 μM
Expand high-hidelity enzyme (3.5 U μl−1)
1
3.5 U
Nuclease-free water
To 50
-
55
Run the PCR as follows:
Table 15
Cycle number
Denature
Anneal
Extend
1
94 °C, 30 s
2–30
94 °C, 30 s
57 °C, 30 s
72 °C, 1 min
31
72 °C, 5 min
-
56
Dilute tenfold by mixing 1 μl of the PCR reaction from Step 55 and 9 μl of H2O. Set up the second PCR reaction as tabulated below:
Table 16
Component
Amount (μl)
Final
Tenfold-diluted DNA from Step 55
2
Unknown
Expand high-fidelity buffer (10×) with MgCl2
5
1×
dNTP (10 mM) mixture
1.5
0.3 mM
Primer Ap2 (50 μM)
1
1 μM
Primer 150L-out or 100R-out (50 μM)
1
1 μM
Expand high-fidelity enzyme (3.5 U μl−1)
1
3.5 U
Nuclease-free water
To 50
-
57
Run the PCR reaction with the same conditions described in Step 55.
-
58
Analyze 10 μl of the sample on a 1.5% (wt/vol) agarose/TAE gel electrophoresis. Purify the DNA bands that are observed in the BAC transgenic fish but not in wild-type control or UASGFP fish from the gel with the QIAquick gel extraction kit according to the manufacturer's instructions. Final volume will be ∼40 μl.
-
59
Use 4 μl of the DNA sample for sequencing with 100L-out, 100R-out and the BigDye terminator cycle sequencing kit according to the manufacturer's instructions. Mix the sample with a one-tenth volume of 3 M sodium acetate and three volumes of 99.6% (vol/vol) ethanol. Incubate the sample at 25 °C for 5 min, centrifuge at 19,300g for 20 min at 4 °C, rinse with 70% (vol/vol) ethanol, dry and suspend in 20 μl of HiDi formamide. Analyze the sample by sequencing (e.g., ABI PRISM 3130xl, Applied Biosystems).
-
60
BLAST the retrieved DNA sequences against the zebrafish genome using Ensembl and NCBI to find the precise location of the Tol2-BAC integration in the genome (Fig. 8c,d).
Troubleshooting
Troubleshooting advice can be found in Table 5. Specific troubleshooting tips for recombineering are available at http://recombineering.ncifcrf.gov/faq.asp.
Timing
Steps 1–5, Identification of BAC clones: 1 d
Steps 6–15, Preparation of BAC-containing competent cells: 4 d
Steps 16–32, Recombineering the iTol2 cassette into the BAC: 2 d
Step 33, Recombineering a reporter gene into the Tol2-BAC plasmid: 2–6 d
Steps 34–44, Microinjection of the Tol2-BAC DNA and TP RNA: 3 d
Steps 45–48, Confirmation of Tol2-mediated BAC excision in injected embryos: 2 d
Step 49, Selection and rearing of Tol2-BAC injected fish: 3–4 months
Step 50, Screening of Tol2-BAC injected fish: 3 d
Steps 51–60, Analysis of Tol2-BAC integrations (optional): 3 d
Box 1, Preparation of SW102 electrocompetent cells: 1 d
Box 2, BAC DNA miniprep: 2 h
Box 3, Preparation of genomic DNA from zebrafish tail fin: 1 d
Anticipated results
Identification of BACs and construction of Tol2-compatible BAC transgenes by recombineering is relatively straightforward (Steps 1–33). It is easy to confirm the success of BAC modifications at every step of the procedure (Fig. 5). To confirm that the Tol2-BAC plasmid is intact and that the Tol2 mRNA works properly during microinjection, we highly recommend carrying out a PCR excision assay (Fig. 4b). After microinjection of 100 or so embryos per construct (Step 44), one can expect that approximately 5–20 of the injected fish will carry a stable germline integration of the Tol2-BAC construct. Germline transmission occurs in ∼15% of injected fish regardless of the source or size of the BAC DNA (70–165 kb and two different libraries, Fig. 7 and Supplementary Fig. 1). Although it is possible that there is a size limit for Tol2 inserts, we have not observed this yet. In the exceptional case where a BAC does not integrate stably in the germline, we assume that sequences in the BAC DNA itself could be unstable or have deleterious effects. Stable BAC transgenic fish are easily identified by detection of a fluorescent reporter inside the BAC (Fig. 7) or by PCR. We can anticipate that the integration site of the BAC construct in the genome can be easily deduced by PCR methods (Fig. 8c,d). Finally, our iTol2 cassettes and protocols for BAC transgenesis are readily applicable to a variety of model organisms, including mice33.
References
Giraldo, P. & Montoliu, L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic. Res. 10, 83–103 (2001).
Nagy, A., Gertsenstein, M., Vintersten, K. & Behringer, R. Manipulating the Mouse Embryo: A Laboratory Manual, 3rd edn. (Cold Spring Harbor Press, Cold Spring Harbor, New York, USA, 2001).
Vintersten, K., Testa, G., Naumann, R., Anastassiadis, K. & Stewart, A.F. Bacterial artificial chromosome transgenesis through pronuclear injection of fertilized mouse oocytes. Methods Mol. Biol. 415, 83–100 (2008).
Copeland, N.G., Jenkins, N.A. & Court, D.L. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2, 769–779 (2001).
Heintz, N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nat. Rev. Neurosci. 2, 861–870 (2001).
Jessen, J.R., Willett, C.E. & Lin, S. Artificial chromosome transgenesis reveals long-distance negative regulation of rag1 in zebrafish. Nat. Genet. 23, 15–16 (1999).
Yang, Z. et al. Modified bacterial artificial chromosomes for zebrafish transgenesis. Methods 39, 183–188 (2006).
Higashijima, S.-I. Transgenic zebrafish expressing fluorescent proteins in central nervous system neurons. Dev. Growth Diff. 50, 407–413 (2008).
Lieschke, G.J. & Currie, P.D. Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 8, 353–367 (2007).
Asakawa, K. et al. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. USA 105, 1255–1260 (2008).
Scott, E. et al. Targeting neural circuitry in zebrafish using Gal4 enhancer trapping. Nat. Methods 4, 323–326 (2007).
Emelyanov, A. & Parinov, S. Mifepristone-inducible LexPR system to drive and control gene expression in transgenic zebrafish. Dev. Biol. 320, 113–121 (2008).
Langenau, D.M. et al. Cre/lox-regulated zebrafish model with conditional myc-induced T-cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 102, 6068–6073 (2005).
Knopf, F. et al. Dually inducible TetON systems for tissue-specific conditional gene expression in zebrafish. Proc. Natl. Acad. Sci. USA 107, 19933–19938 (2010).
Chandler, K.J. et al. Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm. Genome 18, 693–708 (2007).
Garrick, D., Fiering, S., Martin, D.I. & Whitelaw, E. Repeat-induced gene silencing in mammals. Nat. Genet. 18, 56–59 (1998).
Dorer, D.R. Do transgene arrays form heterochromatin in vertebrates? Transgenic. Res. 6, 3–10 (1997).
Stuart, G.W., McMurray, J.V. & Westerfield, M. Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development 103, 403–412 (1988).
Amsterdam, A., Lin, S. & Hopkins, N. The Aequorea victoria green fluorescent protein can be used as a reporter in live zebrafish embryos. Dev. Biol. 171, 123–129 (1995).
Long, Q. et al. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development 124, 4105–4111 (1997).
Higashijima, S., Okamoto, H., Ueno, N., Hotta, Y. & Eguchi, G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev. Biol. 192, 289–299 (1997).
Rembold, M., Lahiri, K., Foulkes, N.S. & Wittbrodt, J. Transgenesis in fish: efficient selection of transgenic fish by co-injection with a fluorescent reporter construct. Nat. Protoc. 1, 1133–1139 (2006).
Lin, S. et al. Integration and germ-line transmission of a pseudotyped retroviral vector in zebrafish. Science 265, 666–669 (1994).
Gaiano, N., Allende, M., Amsterdam, A., Kawakami, K. & Hopkins, N. Highly efficient germ-line transmission of proviral insertions in zebrafish. Proc. Natl. Acad. Sci. USA 93, 7777–7782 (1996).
Ellingsen, S. et al. Large-scale enhancer detection in the zebrafish genome. Development 132, 3799–811 (2005).
Wang, D. et al. Efficient genome-wide mutagenesis of zebrafish genes by retroviral insertions. Proc. Natl. Acad. Sci. USA 104, 12428–12433 (2007).
Thomas, C.E., Ehrhardt, A. & Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 4, 346–358 (2003).
Fadool, J.M., Hartl, D.L. & Dowling, J.E. Transposition of the mariner element from Drosophila mauritiana in zebrafish. Proc. Natl. Acad. Sci. USA 95, 5182–516 (1998).
Kawakami, K. et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7, 133–144 (2004).
Davidson, A.E. et al. Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev. Biol. 263, 191–202 (2003).
Kawakami, K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 8 (Suppl 1): S7 (2007).
Korzh, V. Transposons as tools for enhancer trap screens in vertebrates. Genome Biol. 8 (Suppl 1): S8 (2007).
Suster, M.L., Sumiyama, K. & Kawakami, K. Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genomics 10, 477 (2009).
Kawakami, K. & Shima, A. Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio. Gene 240, 239–244 (1999).
Balciunas, D. et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2, e169 (2006).
Urasaki, A., Morvan, G. & Kawakami, K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174, 639–649 (2006).
Suster, M.L. et al. A novel conserved evx1 enhancer links spinal interneuron morphology and cis-regulation from fish to mammals. Dev. Biol. 325, 422–433 (2009).
Sharan, K., Thomason, L.C., Kuznetsov, S.G. & Court, D.L. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206–223 (2009).
Zhang, Z.D. et al. Statistical analysis of the genomic distribution and correlation of regulatory elements in the ENCODE regions. Genome Res. 17, 787–797 (2007).
Murphy, K.C. Use of bacteriophage l recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180, 2063–2071 (1998).
Murphy, K.C., Campellone, K.G. & Poteete, A.R. PCR-mediated gene replacement in Escherichia coli. Gene 246, 321–330 (2000).
Zhang, Y., Buchholz, F., Muyrers, J.P. & Stewart, A.F. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20, 123–128 (1998).
Zhang, Y., Muyrers, J.P., Testa, G. & Stewart, A.F. DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol. 18, 1314–1317 (2000).
Yu, D. et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97, 5978–5983 (2000).
Warming, S., Costantino, N., Court, D.L., Jenkins, N.A. & Copeland, N.G. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33, e36 (2005).
Cassuto, E. & Radding, C.M. Mechanism for the action of l exonuclease in genetic recombination. Nat. New Biol. 229, 13–16 (1971).
Little, J.W. An exonuclease induced by bacteriophage l. II. Nature of the enzymatic reaction. J. Biol. Chem. 242, 679–686 (1967).
Carter, D.M. & Radding, C.M. The role of exonuclease and b protein of phage l in genetic recombination. II. Substrate specificity and the mode of action of lambda exonuclease. J. Biol. Chem. 246, 2502–2512 (1971).
Cassuto, E., Lash, T., Sriprakash, K.S. & Radding, C.M. Role of exonuclease and b protein of phage l in genetic recombination. V. Recombination of l DNA in vitro. Proc. Natl. Acad. Sci. USA 68, 1639–1643 (1971).
Sadowski, I., Ma, J., Triezenberg, S. & Ptashne, M. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335, 563–564 (1988).
Köster, R.W. & Fraser, S.E. Tracing transgene expression in living zebrafish embryos. Dev. Biol. 233, 329–346 (2001).
Sternberg, N. & Hamilton, D. Bacteriophage P1 site-specific recombination: I. Recombination between loxP sites. J. Mol. Biol. 150, 467–486 (1981).
Suster, M.L., Kikuta, H., Urasaki, A., Asakawa, K. & Kawakami, K. Transgenesis in zebrafish with the tol2 transposon system. Methods Mol. Biol. 561, 41–63 (2009).
Baier, H. & Scott, E.K. Genetic and optical targeting of neural circuits and behavior—zebrafish in the spotlight. Curr. Opin. Neurobiol. 19, 553–560.
Friedenreich, H. & Schartl, M. Transient expression directed by homologous and heterologous promoter and enhancer sequences in fish cells. Nucleic Acids Res. 18, 3299–3305 (1990).
Datta, S., Costantino, N. & Court, D.L. A set of recombineering plasmids for Gram-negative bacteria. Gene 379, 109–115 (2006).
Lister, J.A., Robertson, C.P., Lepage, T., Johnson, S.L. & Raible, D.W. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 126, 3757–3767 (1999).
Acknowledgements
For excellent fish care, we thank T. Kronstad, F. Silva and J. Xu at the Sars International Centre, and A. Ito, M. Suzuki, M. Mizushina, N. Mouri and T. Uematsu at the National Institute of Genetics. We thank A. Urasaki for initial characterization of zT2TP and K. Sumiyama for the pBSK-GFP-pA-FRT-kan-FRT plasmid. This work was supported by core funding from the Sars International Centre, a fellowship from the Japan Society for the Promotion of Science and the Takeda Science Foundation to M.L.S., and grants from the National BioResource Project of Japan and the Ministry of Education, Culture, Sports, Science and Technnology of Japan to K.K.
AUTHOR CONTRIBUTIONS
M.L.S. wrote the manuscript, directed the work at the Sars Centre and generated data and reagents. A.S. contributed reagents and optimized extensively the protocol. G.A. contributed data. K.K directed the work and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Data
Full sequence and graphic map of plasmids used. (PDF 550 kb)
Supplementary Fig. 1
(TIFF 28336 kb)
Rights and permissions
About this article
Cite this article
Suster, M., Abe, G., Schouw, A. et al. Transposon-mediated BAC transgenesis in zebrafish. Nat Protoc 6, 1998–2021 (2011). https://doi.org/10.1038/nprot.2011.416
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.416
This article is cited by
-
Alcam-a and Pdgfr-α are essential for the development of sclerotome-derived stromal cells that support hematopoiesis
Nature Communications (2023)
-
The little skate genome and the evolutionary emergence of wing-like fins
Nature (2023)
-
A53T mutant α-synuclein fibrils formed in macrophage are spread to neurons
Cellular and Molecular Life Sciences (2022)
-
Bacterial Artificial Chromosome-based Protein Expression Platform Using the Tol2 Transposon System
Biotechnology and Bioprocess Engineering (2022)
-
Rapid and Efficient BAC Recombineering: Gain & Loss Screening System
Biotechnology and Bioprocess Engineering (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.