Abstract
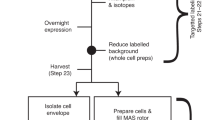
The cell is a crowded environment in which proteins interact specifically with other proteins, nucleic acids, cofactors and ligands. Atomic resolution structural explanation of proteins functioning in this environment is a main goal of biochemical research. Recent improvements to nuclear magnetic resonance (NMR) hardware and methodology allow the measurement of high-resolution heteronuclear multidimensional NMR spectra of macromolecules in living cells (in-cell NMR). In this study, we describe a protocol for the stable isotope (13C, 15N and 2H) labeling and structure determination of proteins overexpressed in Escherichia coli cells exclusively on the basis of information obtained in living cells. The protocol combines the preparation of the protein in E. coli cells, the rapid measurement of the three-dimensional (3D) NMR spectra by nonlinear sampling of the indirectly acquired dimensions, structure calculation and structure refinement. Under favorable circumstances, this in-cell NMR approach can provide high-resolution 3D structures of proteins in living environments. The protocol has been used to solve the first 3D structure of a protein in living cells for the putative heavy metal-binding protein TTHA1718 from Thermus thermophilus HB8 overexpressed in E. coli cells. As no protein purification is necessary, a sample for in-cell NMR measurements can be obtained within 2–3 d. With the nonlinear sampling scheme, the duration of each 3D experiment can be reduced to 2–3 h. Once chemical shift assignments and NOESY peak lists have been prepared, structure calculation with the program CYANA and energy refinement can be completed in less than 1 h on a powerful computer system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ellis, R.J. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 26, 597–604 (2001).
Pielak, G.J. et al. Protein nuclear magnetic resonance under physiological conditions. Biochemistry 48, 226–234 (2009).
Sakakibara, D. et al. Protein structure determination in living cells by in-cell NMR spectroscopy. Nature 458, 102–105 (2009).
Serber, Z. & Dotsch, V. In-cell NMR spectroscopy. Biochemistry 40, 14317–14323 (2001).
Serber, Z., Corsini, L., Durst, F. & Dötsch, V. In-cell NMR spectroscopy. Method. Enzymol. 394, 17–41 (2005).
Serber, Z. et al. Investigating macromolecules inside cultured and injected cells by in-cell NMR spectroscopy. Nat. Protoc. 1, 2701–2709 (2006).
Burz, D.S., Dutta, K., Cowburn, D. & Shekhtman, A. Mapping structural interactions using in-cell NMR spectroscopy (STINT-NMR). Nat. Methods 3, 91–93 (2006).
Burz, D.S., Dutta, K., Cowburn, D. & Shekhtman, A. In-cell NMR for protein-protein interactions (STINT-NMR). Nat. Protoc. 1, 146–152 (2006).
Reckel, S., Hänsel, R., Löhr, F. & Dötsch, V. In-cell NMR spectroscopy. Prog. Nucl. Mag. Res. Sp. 51, 91–101 (2007).
Selenko, P. & Wagner, G. Looking into live cells with in-cell NMR spectroscopy. J. Struct. Biol. 158, 244–253 (2007).
Serber, Z. et al. High-resolution macromolecular NMR spectroscopy inside living cells. J. Am. Chem. Soc. 123, 2446–2447 (2001).
Serber, Z., Ledwidge, R., Miller, S.M. & Dötsch, V. Evaluation of parameters critical to observing proteins inside living Escherichia coli by in-cell NMR spectroscopy. J. Am. Chem. Soc. 123, 8895–8901 (2001).
Wieruszeski, J.M., Bohin, A., Bohin, J.P. & Lippens, G. In vivo detection of the cyclic osmoregulated periplasmic glucan of Ralstonia solanacearum by high-resolution magic angle spinning NMR. J. Magn. Reson. 151, 118–123 (2001).
Selenko, P., Serber, Z., Gade, B., Ruderman, J. & Wagner, G. Quantitative NMR analysis of the protein G B1 domain in Xenopus laevis egg extracts and intact oocytes. Proc. Natl. Acad. Sci. USA 103, 11904–11909 (2006).
Sakai, T. et al. In-cell NMR spectroscopy of proteins inside Xenopus laevis oocytes. J. Biomol. NMR 36, 179–188 (2006).
Bodart, J.F. et al. NMR observation of Tau in Xenopus oocytes. J. Magn. Reson. 192, 252–257 (2008).
Inomata, K. et al. High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature 458, 106–109 (2009).
Ogino, S. et al. Observation of NMR signals from proteins introduced into living mammalian cells by reversible membrane permeabilization using a pore-forming toxin, streptolysin O. J. Am. Chem. Soc. 131, 10834–10835 (2009).
Barna, J.C.J., Laue, E.D., Mayger, M.R., Skilling, J. & Worrall, S.J.P. Exponential sampling, an alternative method for sampling in two-dimensional NMR experiments. J. Magn. Reson. 73, 69–77 (1987).
Schmieder, P., Stern, A.S., Wagner, G. & Hoch, J.C. Improved resolution in triple-resonance spectra by nonlinear sampling in the constant-time domain. J. Biomol. NMR 4, 483–490 (1994).
Rovnyak, D. et al. Accelerated acquisition of high resolution triple-resonance spectra using non-uniform sampling and maximum entropy reconstruction. J. Magn. Reson. 170, 15–21 (2004).
Rosen, M.K. et al. Selective methyl group protonation of perdeuterated proteins. J. Mol. Biol. 263, 627–636 (1996).
Serber, Z. et al. Methyl groups as probes for proteins and complexes in in-cell NMR experiments. J. Am. Chem. Soc. 126, 7119–7125 (2004).
Güntert, P. Automated NMR protein structure calculation. Prog. Nucl. Mag. Res. Sp. 43, 105–125 (2003).
Williams, S.P., Haggie, P.M. & Brindle, K.M. 19F NMR measurements of the rotational mobility of proteins in vivo. Biophys. J. 72, 490–498 (1997).
Laue, E.D., Mayger, M.R., Skilling, J. & Staunton, J. Reconstruction of phase sensitive 2D NMR spectra by maximum entropy. J. Magn. Reson. 68, 14–29 (1986).
Herrmann, T., Güntert, P. & Wüthrich, K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 (2002).
Güntert, P., Mumenthaler, C. & Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 (1997).
Cornilescu, G., Delaglio, F. & Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 (1999).
Cornell, W.D. et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117, 5179–5197 (1995).
Luginbühl, P., Güntert, P., Billeter, M. & Wüthrich, K. The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J. Biomol. NMR 8, 136–146 (1996).
Koradi, R., Billeter, M. & Güntert, P. Point-centered domain decomposition for parallel molecular dynamics simulation. Comput. Phys. Commun. 124, 139–147 (2000).
Hoch, J.C. & Stern, A.S. NMR Data Processing (Wiley-Liss, New York, 1996).
Kraulis, P.J. ANSIG: a program for the assignment of protein 1H 2D NMR spectra by interactive computer graphics. J. Magn. Reson. 84, 627–633 (1989).
Kraulis, P.J., Domaille, P.J., Campbell-Burk, S.L., Van Aken, T. & Laue, E.D. Solution structure and dynamics of Ras p21-GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry 33, 3515–3531 (1994).
Vranken, W.F. et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 (2005).
Johnson, B.A. & Blevins, R.A. NMR view: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994).
Johnson, B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 (2004).
Bartels, C., Xia, T., Billeter, M., Güntert, P. & Wüthrich, K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 6, 1–10 (1995).
Shen, Y., Delaglio, F., Cornilescu, G. & Bax, A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009).
Jaravine, V.A. & Orekhov, V.Y. Targeted acquisition for real-time NMR spectroscopy. J. Am. Chem. Soc. 128, 13421–13426 (2006).
Jaravine, V., Ibraghimov, I. & Orekhov, V.Y. Removal of a time barrier for high-resolution multidimensional NMR spectroscopy. Nat. Methods 3, 605–607 (2006).
Marion, D. Fast acquisition of NMR spectra using Fourier transform of non-equispaced data. J. Biomol. NMR 32, 141–150 (2005).
Kazimierczuk, K., Zawadzka, A., Kozminski, W. & Zhukov, I. Random sampling of evolution time space and Fourier transform processing. J. Biomol. NMR 36, 157–168 (2006).
Takeda, M., Ikeya, T., Güntert, P. & Kainosho, M. Automated structure determination of proteins with the SAIL-FLYA NMR method. Nat. Protoc. 2, 2896–2902 (2007).
Thorstenson, Y.R., Zhang, Y., Olson, P.S. & Mascarenhas, D. Leaderless polypeptides efficiently extracted from whole cells by osmotic shock. J. Bacteriol. 179, 5333–5339 (1997).
Gardy, J.L. et al. PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 31, 3613–3617 (2003).
Gardy, J.L. et al. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21, 617–623 (2005).
Nielsen, H., Engelbrecht, J., Brunak, S. & von Heijne, G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6 (1997).
Bendtsen, J.D., Nielsen, H., von Heijne, G. & Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 (2004).
Li, C. et al. Differential dynamical effects of macromolecular crowding on an intrinsically disordered protein and a globular protein: implications for in-cell NMR spectroscopy. J. Am. Chem. Soc. 130, 6310–6311 (2008).
Acknowledgements
We thank Professor Seiki Kuramitsu for providing the plasmid encoding TTHA1718. This work was supported in part by the CREST program of the Japan Science and Technology Agency (JST), the Molecular Ensemble Program of RIKEN, Grants-in-Aid for Scientific Research of Priority Areas from the Japanese Ministry of Education, Sports, Culture, Science, and Technology on 'Molecular Soft Interactions Regulating Membrane Interface of Biological Systems' and 'Molecular Science for Supra Functional Systems—Development of Advanced Methods for Exploring Elementary Process', and by the Volkswagen Foundation.
Author information
Authors and Affiliations
Contributions
B.O.S., M.S., P.G. and Y.I. designed the research and wrote the article. T.I. developed the protocol for structure calculation and refinement. A.S., D.S., J.H., T.H., M.Y., N.H. and T.M. developed the protocols, including sample preparation and characterization, data acquisition and resonance assignment. Y.S., M.M., D.N. and M.W. contributed the protocol development for rapid NMR data acquisition and MaxEnt processing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Ikeya, T., Sasaki, A., Sakakibara, D. et al. NMR protein structure determination in living E. coli cells using nonlinear sampling. Nat Protoc 5, 1051–1060 (2010). https://doi.org/10.1038/nprot.2010.69
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.69
This article is cited by
-
In-cell NMR as a sensitive tool to monitor physiological condition of Escherichia coli
Scientific Reports (2020)
-
Improved in-cell structure determination of proteins at near-physiological concentration
Scientific Reports (2016)
-
CcpNmr AnalysisAssign: a flexible platform for integrated NMR analysis
Journal of Biomolecular NMR (2016)
-
A new carbamidemethyl-linked lanthanoid chelating tag for PCS NMR spectroscopy of proteins in living HeLa cells
Journal of Biomolecular NMR (2016)
-
In-cell 13C NMR spectroscopy for the study of intrinsically disordered proteins
Nature Protocols (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.