Abstract
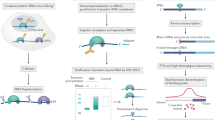
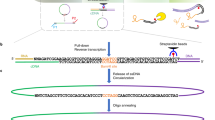
Cross-linking and immunoprecipitation (CLIP) is increasingly used to map transcriptome-wide binding sites of RNA-binding proteins. We developed a method for CLIP data analysis, and applied it to compare CLIP with photoactivatable ribonucleoside–enhanced CLIP (PAR-CLIP) and to uncover how differences in cross-linking and ribonuclease digestion affect the identified sites. We found only small differences in accuracies of these methods in identifying binding sites of HuR, which binds low-complexity sequences, and Argonaute 2, which has a complex binding specificity. We found that cross-link–induced mutations led to single-nucleotide resolution for both PAR-CLIP and CLIP. Our results confirm the expectation from original CLIP publications that RNA-binding proteins do not protect their binding sites sufficiently under the denaturing conditions used during the CLIP procedure, and we show that extensive digestion with sequence-specific RNases strongly biases the recovered binding sites. This bias can be substantially reduced by milder nuclease digestion conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Brown, V. et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107, 477–487 (2001).
Eystathioy, T. et al. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell 13, 1338–1351 (2002).
Landthaler, M. et al. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 14, 2580–2596 (2008).
Mukherjee, N., Lager, P.J., Friedersdorf, M.B., Thompson, M.A. & Keene, J.D. Coordinated posttranscriptional mRNA population dynamics during T-cell activation. Mol. Syst. Biol. 5, 288 (2009).
Keene, J.D. & Tenenbaum, S.A. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9, 1161–1167 (2002).
Bhattacharyya, S.N., Habermacher, R., Martine, U., Closs, E.I. & Filipowicz, W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124 (2006).
Kedde, M. et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131, 1273–1286 (2007).
Kim, H.H. et al. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 23, 1743–1748 (2009).
Kedde, M. et al. A Pumilio-induced RNA structure switch in p27–3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 12, 1014–1020 (2010).
Ule, J. et al. CLIP identifies Nova-regulated RNA networks in the brain. Science 302, 1212–1215 (2003).
Licatalosi, D.D. et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469 (2008).
Hafner, M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010).
Konig, J. et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 17, 909–915 (2010).
Atasoy, U., Watson, J., Patel, D. & Keene, J.D. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J. Cell Sci. 111, 3145–3156 (1998).
Ray, D. et al. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 27, 667–670 (2009).
Ender, C. & Meister, G. Argonaute proteins at a glance. J. Cell Sci. 123, 1819–1823 (2010).
Ule, J., Jensen, K., Mele, A. & Darnell, R.B. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 37, 376–386 (2005).
Chi, S.W., Zang, J.B., Mele, A. & Darnell, R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486 (2009).
Wang, Z., Tollervey, J., Briese, M., Turner, D. & Ule, J. CLIP: construction of cDNA libraries for high-throughput sequencing from RNAs cross-linked to proteins in vivo. Methods 48, 287–293 (2009).
Zisoulis, D.G. et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 17, 173–179 (2010).
Takahashi, K., Uchida, T. & Egami, F. Ribonuclease T1, structure and function. Adv. Biophys. 1, 53–98 (1970).
Dingwall, C., Lomonossoff, G.P. & Laskey, R.A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 9, 2659–2673 (1981).
Granneman, S., Kudla, G., Petfalski, E. & Tollervey, D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. USA 106, 9613–9618 (2009).
Wang, X. & Tanaka Hall, T.M. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat. Struct. Biol. 8, 141–145 (2001).
Khorshid, M., Rodak, C. & Zavolan, M. CLIPZ: a database and analysis environment for experimentally determined binding sites of RNA-binding proteins. Nucleic Acids Res. 39, D245–D252 (2011).
Levine, T.D., Gao, F., King, P.H., Andrews, L.G. & Keene, J.D. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol. Cell. Biol. 13, 3494–3504 (1993).
Fan, X.C. & Steitz, J.A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17, 3448–3460 (1998).
Lewis, B.P., Burge, C.B. & Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
Smith, K.C. & Meun, D.H. Kinetics of the photochemical addition of [35S] cysteine to polynucleotides and nucleic acids. Biochemistry 7, 1033–1037 (1968).
Hockensmith, J.W., Kubasek, W.L., Vorachek, W.R. & von Hippel, P.H. Laser cross-linking of nucleic acids to proteins. Methodology and first applications to the phage T4 DNA replication system. J. Biol. Chem. 261, 3512–3518 (1986).
Rudel, S., Flatley, A., Weinmann, L., Kremmer, E. & Meister, G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA 14, 1244–1253 (2008).
Krol, J. et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141, 618–631 (2010).
Berninger, P., Gaidatzis, D., van Nimwegen, E. & Zavolan, M. Computational analysis of small RNA cloning data. Methods 44, 13–21 (2008).
Fraley, C. & Raftery, A.E. Model-based clustering, discriminant analysis, and density estimation. J. Am. Stat. Assoc. 9, 611–631 (2002).
Smyth, G.K. Limma: linear models for microarray data. in Bioinformatics and Computational Biology Solutions using R and Bioconductor (eds., Gentleman, R., Carey, V., Dudoit, S., Irizarry, R. & Huber, W.) 397–420 (Springer, New York, 2005).
Acknowledgements
We thank G. Meister (University of Regensburg) for Ago2 antibody, C. Rammelt for initial help with PAR-CLIP, I. Nissen and C. Beisel for support with deep sequencing, J. Krol for help with miRNA quantification, B. Dimitriades for help with cell culture, and W. Keller and W. Filipowicz for critical comments and suggestions. We also thank the members of the Zavolan laboratory for valuable discussions. This work was supported by grants from the Swiss National Science Foundation (#31003A_127307, PDAM_3_127218 and PDFMP3_123123), the Swiss Initiative in Systems Biology (Cell Plasticity) and the Swiss Cancer League (#KFS 02477-08-2009) and by the European Molecular Biology Organization long-term fellowship 184-2009 to S.K.
Author information
Authors and Affiliations
Contributions
S.K. designed and performed the experiments, L.J. designed and performed the experiments and wrote the paper, L.B. analyzed the data, J.H. analyzed the data, M.K. developed the annotation tools and analyzed the data and M.Z. designed and supervised the experiments, analyzed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14 and Supplementary Table 1 (PDF 10786 kb)
Rights and permissions
About this article
Cite this article
Kishore, S., Jaskiewicz, L., Burger, L. et al. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods 8, 559–564 (2011). https://doi.org/10.1038/nmeth.1608
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1608
This article is cited by
-
Functional role and epithelial to mesenchymal transition of the miR-590-3p/MDM2 axis in hepatocellular carcinoma
BMC Cancer (2023)
-
Multi-omics analysis revealed TEK and AXIN2 are potential biomarkers in multifocal papillary thyroid cancer
Cancer Cell International (2022)
-
Dual function of SF3B2 on chromatin and RNA to regulate transcription in head and neck squamous cell carcinoma
Cell & Bioscience (2022)
-
Competing endogenous RNA network mediated by circ_3205 in SARS-CoV-2 infected cells
Cellular and Molecular Life Sciences (2022)
-
APE1 controls DICER1 expression in NSCLC through miR-33a and miR-130b
Cellular and Molecular Life Sciences (2022)