Abstract
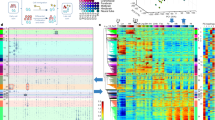
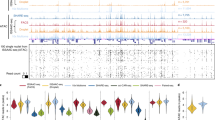
The study of developmentally regulated transcription factors by chromatin immunoprecipitation and deep sequencing (ChIP-seq) faces two major obstacles: availability of ChIP-grade antibodies and access to sufficient number of cells. We describe versatile genome-wide analysis of transcription-factor binding sites by combining directed differentiation of embryonic stem cells and inducible expression of tagged proteins. We demonstrate its utility by mapping DNA-binding sites of transcription factors involved in motor neuron specification.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Park, P.J. Nat. Rev. Genet. 10, 669–680 (2009).
Iacovino, M. et al. Stem Cells 29, 1580–1588 (2011).
Wichterle, H., Lieberam, I., Porter, J.A. & Jessell, T.M. Cell 110, 385–397 (2002).
Novitch, B.G., Chen, A.I. & Jessell, T.M. Neuron 31, 773–789 (2001).
Mukouyama, Y.S. et al. Proc. Natl. Acad. Sci. USA 103, 1551–1556 (2006).
Chen, J.A. et al. Neuron 69, 721–735 (2011).
Jessell, T.M. Nat. Rev. Genet. 1, 20–29 (2000).
Guo, Y. et al. Bioinformatics 26, 3028–3034 (2010).
Jung, H. et al. Neuron 67, 781–796 (2010).
Kim, J., Cantor, A.B., Orkin, S.H. & Wang, J. Nat. Protoc. 4, 506–517 (2009).
Wang, J., Cantor, A.B. & Orkin, S.H. Curr. Protoc. Stem Cell Biol. published online 1 March 2009 (doi: 10.1002/9780470151808.sc01b05s8). (2009).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Genome Biol. 10, R25 (2009).
Benjamini, Y. & Hochberg, Y. J. Royal Stat. Soc. B 57, 289–300 (1995).
van Heeringen, S.J. & Veenstra, G.J. Bioinformatics 27, 270–271 (2011).
Mahony, S. & Benos, P.V. Nucleic. Acids. Res. 35, W253–W258 (2007).
Acknowledgements
E.O.M. is funded as the David and Sylvia Lieb Fellow of the Damon Runyon Cancer Research Foundation (DRG-1937-07). We thank R. Sherwood (Harvard University) for sharing unpublished observations and T Jessell (Columbia University) for anti-Hoxc4 antibody. Personnel and work were supported by US National Institutes of Health grant P01 NS055923 (D.K.G., R.A.Y. and H.W.), R01 NS058502 (H.W.), R01HL081186 (M.K.) and Helmsley Stem Cell Starter grant (H.W.).
Author information
Authors and Affiliations
Contributions
E.O.M. and G.M. generated the transcription factor inducible lines; E.O.M. performed phenotypic analysis of the derived lines. E.O.M., W.A.W. and C.A.M. performed ChIP experiments. M.I. and M.K. developed the ICE cell lines and vectors. E.O.M. performed expression analysis. E.O.M. and M.C. performed the western blot and protein-binding to immobilized DNA. S.M. analyzed ChIP-seq data. E.O.M., R.A.Y., D.K.G .and H.W. designed the experiments. E.O.M., S.M. and H.W. wrote the manuscript; D.K.G. revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Table 1 (PDF 1439 kb)
Rights and permissions
About this article
Cite this article
Mazzoni, E., Mahony, S., Iacovino, M. et al. Embryonic stem cell–based mapping of developmental transcriptional programs. Nat Methods 8, 1056–1058 (2011). https://doi.org/10.1038/nmeth.1775
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1775
This article is cited by
-
An interpretable bimodal neural network characterizes the sequence and preexisting chromatin predictors of induced transcription factor binding
Genome Biology (2021)
-
Vsx1 and Chx10 paralogs sequentially secure V2 interneuron identity during spinal cord development
Cellular and Molecular Life Sciences (2020)
-
Proneural factors Ascl1 and Neurog2 contribute to neuronal subtype identities by establishing distinct chromatin landscapes
Nature Neuroscience (2019)
-
Optical High Content Nanoscopy of Epigenetic Marks Decodes Phenotypic Divergence in Stem Cells
Scientific Reports (2017)
-
Internal epitope tagging informed by relative lack of sequence conservation
Scientific Reports (2016)