Abstract
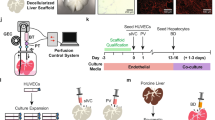
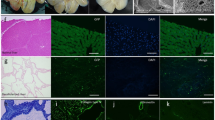
Orthotopic liver transplantation is the only available treatment for severe liver failure, but it is currently limited by organ shortage. One technical challenge that has thus far limited the development of a tissue-engineered liver graft is oxygen and nutrient transport. Here we demonstrate a novel approach to generate transplantable liver grafts using decellularized liver matrix. The decellularization process preserves the structural and functional characteristics of the native microvascular network, allowing efficient recellularization of the liver matrix with adult hepatocytes and subsequent perfusion for in vitro culture. The recellularized graft supports liver-specific function including albumin secretion, urea synthesis and cytochrome P450 expression at comparable levels to normal liver in vitro. The recellularized liver grafts can be transplanted into rats, supporting hepatocyte survival and function with minimal ischemic damage. These results provide a proof of principle for the generation of a transplantable liver graft as a potential treatment for liver disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Heron, M. et al. Deaths: Final Data for 2006. in National Vital Statistics Report, Vol. 57 (Centers for Disease Control and Prevention, Hyattsville, Maryland, 2009).
Punch, J.D., Hayes, D.H., LaPorte, F.B., McBride, V. & Seely, M.S. Organ donation and utilization in the United States, 1996–2005. Am. J. Transplant. 7, 1327–1338 (2007).
Fox, I.J. & Roy-Chowdhury, J. Hepatocyte transplantation. J. Hepatol. 40, 878–886 (2004).
Strom, S.C. et al. Hepatocyte transplantation as a bridge to orthotopic liver transplantation in terminal liver failure. Transplantation 63, 559–569 (1997).
Horslen, S.P. et al. Isolated hepatocyte transplantation in an infant with a severe urea cycle disorder. Pediatrics 111, 1262–1267 (2003).
Fox, I.J. et al. Treatment of the Crigler-Najjar syndrome type I with hepatocyte transplantation. N. Engl. J. Med. 338, 1422–1426 (1998).
Fisher, R.A. & Strom, S.C. Human hepatocyte transplantation: worldwide results. Transplantation 82, 441–449 (2006).
Habibullah, C.M., Syed, I.H., Qamar, A. & Taher-Uz, Z. Human fetal hepatocyte transplantation in patients with fulminant hepatic failure. Transplantation 58, 951–952 (1994).
Nagata, H. et al. Prolonged survival of porcine hepatocytes in cynomolgus monkeys. Gastroenterology 132, 321–329 (2007).
Basma, H. et al. Differentiation and transplantation of human embryonic stem cell–derived hepatocytes. Gastroenterology 136, 990–999 (2009).
Cho, C.H. et al. Homogeneous differentiation of hepatocyte-like cells from embryonic stem cells: applications for the treatment of liver failure. FASEB J. 22, 898–909 (2008).
Totsugawa, T. et al. Survival of liver failure pigs by transplantation of reversibly immortalized human hepatocytes with tamoxifen-mediated self-recombination. J. Hepatol. 47, 74–82 (2007).
Shafritz, D.A., Oertel, M., Menthena, A., Nierhoff, D. & Dabeva, M.D. Liver stem cells and prospects for liver reconstitution by transplanted cells. Hepatology 43, S89–S98 (2006).
Smets, F., Najimi, M. & Sokal, E.M. Cell transplantation in the treatment of liver diseases. Pediatr. Transplant. 12, 6–13 (2008).
Langer, R. & Vacanti, J.P. Tissue engineering. Science 260, 920–926 (1993).
Wang, S., Nagrath, D., Chen, P.C., Berthiaume, F. & Yarmush, M.L. Three-dimensional primary hepatocyte culture in synthetic self-assembling peptide hydrogel. Tissue. Eng. Part A 14, 227–236 (2008).
Nahmias, Y., Berthiaume, F. & Yarmush, M.L. Integration of technologies for hepatic tissue engineering. Adv. Biochem. Eng. Biotechnol. 103, 309–329 (2007).
Ohashi, K. et al. Engineering functional two- and three-dimensional liver systems in vivo using hepatic tissue sheets. Nat. Med. 13, 880–885 (2007).
Kulig, K.M. & Vacanti, J.P. Hepatic tissue engineering. Transpl. Immunol. 12, 303–310 (2004).
Nahmias, Y., Schwartz, R.E., Hu, W.S., Verfaillie, C.M. & Odde, D.J. Endothelium-mediated hepatocyte recruitment in the establishment of liver-like tissue in vitro. Tissue Eng. 12, 1627–1638 (2006).
Badylak, S.F. The extracellular matrix as a biologic scaffold material. Biomaterials 28, 3587–3593 (2007).
Yoo, J.J., Meng, J., Oberpenning, F. & Atala, A. Bladder augmentation using allogenic bladder submucosa seeded with cells. Urology 51, 221–225 (1998).
Dahl, S.L., Koh, J., Prabhakar, V. & Niklason, L.E. Decellularized native and engineered arterial scaffolds for transplantation. Cell Transplant. 12, 659–666 (2003).
Nieponice, A., Gilbert, T.W. & Badylak, S.F. Reinforcement of esophageal anastomoses with an extracellular matrix scaffold in a canine model. Ann. Thorac. Surg. 82, 2050–2058 (2006).
Schechner, J.S. et al. Engraftment of a vascularized human skin equivalent. FASEB J. 17, 2250–2256 (2003).
Macchiarini, P. et al. Clinical transplantation of a tissue-engineered airway. Lancet 372, 2023–2030 (2008).
Ott, H.C. et al. Perfusion-decellularized matrix: using nature's platform to engineer a bioartificial heart. Nat. Med. 14, 213–221 (2008).
Safar, P. Clinical death symposium. Crit. Care Med. 16, 919–920 (1988).
Hoffenberg, R. Measurement of the synthesis of liver-produced plasma proteins with particular reference to dietary protein and amino acid supply. Biochem. J. 129, 3P (1972).
Khetani, S.R. & Bhatia, S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 26, 120–126 (2008).
Tolboom, H. et al. Recovery of warm ischemic rat liver grafts by normothermic extracorporeal perfusion. Transplantation 87, 170–177 (2009).
Uygun, K. et al. Diluted blood reperfusion as a model for transplantation of ischemic rat livers: alt is a direct indicator of viability. Transplant. Proc. (in the press).
Cho, C.H. et al. Oxygen uptake rates and liver-specific functions of hepatocyte and 3T3 fibroblast co-cultures. Biotechnol. Bioeng. 97, 188–199 (2007).
Rotem, A. et al. Oxygen is a factor determining in vitro tissue assembly: effects on attachment and spreading of hepatocytes. Biotechnol. Bioeng. 43, 654–660 (1994).
Foy, B.D. et al. Optimization of hepatocyte attachment to microcarriers: importance of oxygen. Biotechnol. Bioeng. 42, 579–588 (1993).
Balis, U.J. et al. Oxygen consumption characteristics of porcine hepatocytes. Metab. Eng. 1, 49–62 (1999).
Dunn, J.C., Tompkins, R.G. & Yarmush, M.L. Long-term in vitro function of adult hepatocytes in a collagen sandwich configuration. Biotechnol. Prog. 7, 237–245 (1991).
Schleimer, K. et al. Improved technique of heterotopic auxiliary rat liver transplantation with portal vein arterialization. Langenbecks Arch. Surg. 391, 102–107 (2006).
Acknowledgements
This work was supported by grants from the US National Institutes of Health, R01DK59766 and R01DK084053 to M.L.Y., K99/R00 DK080942 to K.U. and K99DK083556 to A.S.-G., and US National Science Foundation CBET-0853569 to K.U. We thank the support of the Shriners Hospitals for Children to B.E.U. (grant no. 8503), the American Liver Foundation to A.S.-G. and Massachusetts General Hospital Junior Faculty Grant to K.U. and the Shriners Hospitals for Children. We would like to thank A. Vitalo, C. Calhoun and B. Crowther for technical support.
Author information
Authors and Affiliations
Contributions
K.U. had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analyses. B.E.U., A.S.-G., H.Y. and K.U., study concept and design; B.E.U., A.S.-G., H.Y., M.A.G., acquisition of data; B.E.U., A.S.-G., Y.N. and K.U., analysis and interpretation of data; M.-L.I., C.S. and B.E.U., rat liver harvest, decellularization and hepatocyte isolation; J.M. and B.E.U., design and construction of the recellularized liver chamber; B.E.U., A.S.-G., H.Y. and M.A.G., recellularization; A.S.-G. and H.Y., histology and transplantation studies; B.E.U., A.S.-G., H.Y. and K.U., drafting of the manuscript; B.E.U., A.S.-G., H.Y., M.L.Y., Y.N., A.T., F.B., M.H., N.K. and K.U., critical revision of the manuscript for intellectual content; B.E.U. and K.U., statistical analysis; M.L.Y., K.U. and A.S.-G., obtained funding; M.-L.I., C.S., J.M., Y.N., A.T. and F.B., administrative, technical or material support. All authors contributed to the preparation of the report.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1 and 2, Supplementary Figures 1–5 and Supplementary Methods (PDF 4275 kb)
Rights and permissions
About this article
Cite this article
Uygun, B., Soto-Gutierrez, A., Yagi, H. et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med 16, 814–820 (2010). https://doi.org/10.1038/nm.2170
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2170
This article is cited by
-
Bioengineering of vascularized porcine flaps using perfusion-recellularization
Scientific Reports (2024)
-
Three Dimensional Bioprinting for Hepatic Tissue Engineering: From In Vitro Models to Clinical Applications
Tissue Engineering and Regenerative Medicine (2024)
-
Biomimetic natural biomaterials for tissue engineering and regenerative medicine: new biosynthesis methods, recent advances, and emerging applications
Military Medical Research (2023)
-
Effect of different decellularization protocols on reendothelialization with human cells for a perfused renal bioscaffold of the rat
BMC Biotechnology (2023)
-
Nano-biomaterials and advanced fabrication techniques for engineering skeletal muscle tissue constructs in regenerative medicine
Nano Convergence (2023)