Abstract
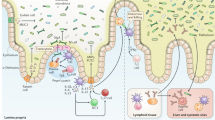
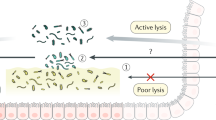
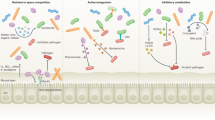
The gut contains our largest collection of resident microorganisms. One obvious question is how microbial communities establish and maintain themselves within a perfused intestine. The answers, which may come in part from observations made by environmental engineers and glycobiologists, have important implications for immunologists who wish to understand how indigenous microbial communities are accommodated. Here we propose that the mucus gel layer overlying the intestinal epithelium is a key contributor to the structural and functional stability of this microbiota and its tolerance by the host.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Savage, D.C. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31, 107–133 (1977).
Hooper, L.V., Midtvedt, T. & Gordon, J.I. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22, 283–307 (2002).
Zoetendal, E.G., Akkermans, A.D., Akkermans-van Vliet, W.M., de Visser, J.A.G.M. & de Vos, W.M. The host genotype affects the bacterial community in the human gastronintestinal tract. Microb. Eco. Health Dis. 13, 129–134 (2001).
Hayashi, H., Sakamoto, M. & Benno, Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46, 535–548 (2002).
Eckburg, P.B., Lepp, P.W. & Relman, D.A. Archaea and their potential role in human disease. Infect. Immun. 71, 591–596 (2003).
Breitbart, M. et al. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185, 6220–6223 (2003).
Stahl, D.A. & Tiedje, J. Microbial ecology and genomics: a crossroads of opportunity, critical issues colloquia. Am. Soc. Microbiol. (Washington, DC, 2002).
Buckley, M.R. Microbial communities: from life apart to life together, critical issues colloquia. Am. Soc. Microbiol. (Washington, DC, 2003).
Suau, A. et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65, 4799–4807 (1999).
Xu, J. & Gordon, J.I. Inaugural article: honor thy symbionts. Proc. Natl. Acad. Sci. USA 100, 10452–10459 (2003).
Venter, J.C. et al. Environmental genome shotgun sequencing of the sargasso sea. Science advance online publication, 4 March 2004 (doi:10.1126/science.1093857).
Bouma, G. & Strober, W. The immunological and genetic basis of inflammatory bowel disease. Nat. Rev. Immunol. 3, 521–533 (2003).
Schembri, M.A., Kjaergaard, K. & Klemm, P. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48, 253–267 (2003).
Beloin, C. et al. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51, 659–674 (2004).
Jensen, E.T., Kharazmi, A., Hoiby, N. & Costerton, J.W. Some bacterial parameters influencing the neutrophil oxidative burst response to Pseudomonas aeruginosa biofilms. APMIS 100, 727–733 (1992).
Jensen, E.T. et al. Complement activation by Pseudomonas aeruginosa biofilms. Microb. Pathog. 15, 377–388 (1993).
Meluleni, G.J., Grout, M., Evans, D.J. & Pier, G.B. Mucoid Pseudomonas aeruginosa growing in a biofilm in vitro are killed by opsonic antibodies to the mucoid exopolysaccharide capsule but not by antibodies produced during chronic lung infection in cystic fibrosis patients. J. Immunol. 155, 2029–2038 (1995).
Rescigno, M. et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2, 361–367 (2001).
Hooper, L.V., Stappenbeck, T.S., Hong, C.V. & Gordon, J.I. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 4, 269–273 (2003).
Kelly, D. et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat. Immunol. 5, 104–112 (2004).
Suzuki, K. et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. USA 101, 1981–1986 (2004).
Fagarasan, S. et al. In situ class switching and differentiation to IgA-producing cells in the gut lamina propria. Nature 413, 639–643 (2001).
Macpherson, A.J. & Uhr, T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303, 1662–1665 (2004).
Treiner, E. et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 (2003).
Shroff, K.E., Meslin, K. & Cebra, J.J. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immun. 63, 3904–3913 (1995).
Bollinger, R.R. et al. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology 109, 580–587 (2003).
Sarti, A., Vieira, L.G., Foresti, E. & Zaiat, M. Influence of the liquid-phase mass transfer on the performance of a packed-bed bioreactor for wastewater treatment. Bioresour. Technol. 78, 231–238 (2001).
Rittmann, B.E. & McCarty, P.L. Environmental Biotechnology (McGraw Hill, Boston, 2000).
Molly, K., Vande, Woestyne, M. & Verstraete, W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 39, 254–258 (1993).
De Boever, P., Deplancke, B. & Verstraete, W. Fermentation by gut microbiota cultured in a simulator of the human intestinal microbial ecosystem is improved by supplementing a soygerm powder. J. Nutr. 130, 2599–2606 (2000).
Lettinga, G., Van Velsen, A., Hobma, S. & Zeeuw, W. Use of the upflow sludge blanket (USB) reactor concept for biological wastewater treatment, especially for anaerobic treatment. Biotech. Bioeng. 22, 699–734 (1980).
Angenent, L., Sung, S. & Raskin, L. Formation of granules and Methanosaeta fibres in an anaerobic migrating blanket reactor (AMBR). Environ. Microbiol. 6, 315–322 (2004).
Matsuo, K., Ota, H., Akamatsu, T., Sugiyama, A. & Katsuyama, T. Histochemistry of the surface mucous gel layer of the human colon. Gut 40, 782–789 (1997).
Robbe, C. et al. Evidence of regio-specific glycosylation in human intestinal mucins: presence of an acidic gradient along the intestinal tract. J. Biol. Chem. 278, 46337–46348 (2003).
Hooper, L.V., Xu, J., Falk, P.G., Midtvedt, T. & Gordon, J.I. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA 96, 9833–9838 (1999).
Akiyama, Y., Nagahara, N., Kashihara, T., Hirai, S. & Toguchi, H. In vitro and in vivo evaluation of mucoadhesive microspheres prepared for the gastrointestinal tract using polyglycerol esters of fatty acids and a poly(acrylic acid) derivative. Pharm. Res. 12, 397–405 (1995).
Nyholm, S.V. & McFall-Ngai, M.J. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl. Environ. Microbiol. 69, 3932–3937 (2003).
Xu, J. et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076 (2003).
Chang, D.E. et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 101 7427–7432 (2004).
Salyers, A.A., Pajeau, M. & McCarthy, R.E. Importance of mucopolysaccharides as substrates for Bacteroides thetaiotaomicron growing in intestinal tracts of exgermfree mice. Appl. Environ. Microbiol. 54, 1970–1976 (1988).
Hwa, V. & Salyers, A.A. Analysis of two chondroitin sulfate utilization mutants of Bacteroides thetaiotaomicron that differ in their abilities to compete with the wild type in the gastrointestinal tracts of germfree mice. Appl. Environ. Microbiol. 58, 869–876 (1992).
Moller, S. et al. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64, 721–732 (1998).
Itoh, H., Beck, P.L., Inoue, N., Xavier, R. & Podolsky, D.K. A paradoxical reduction in susceptibility to colonic injury upon targeted transgenic ablation of goblet cells. J. Clin. Invest. 104, 1539–1547 (1999).
Schreiber, S., Stuben, M., Josenhans, C., Scheid, P. & Suerbaum, S. In vivo distribution of Helicobacter felis in the gastric mucus of the mouse: experimental method and results. Infect. Immun. 67, 5151–5156 (1999).
Orphan, V.J., House, C.H., Hinrichs, K.U., McKeegan, K.D. & DeLong, E.F. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293, 484–487 (2001).
Krinos, C.M. et al. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414, 555–558 (2001).
Coyne, M.J., Weinacht, K.G., Krinos, C.M. & Comstock, L.E. Mpi recombinase globally modulates the surface architecture of a human commensal bacterium. Proc. Natl. Acad. Sci. USA 100, 10446–10451 (2003).
Macfarlane, S., McBain, A.J. & Macfarlane, G.T. Consequences of biofilm and sessile growth in the large intestine. Adv. Dent. Res. 11, 59–68 (1997).
Probert, H.M. & Gibson, G.R. Bacterial biofilms in the human gastrointestinal tract. Curr. Issues Intest. Microbiol. 3, 23–27 (2002).
Mahan, M.J., Slauch, J.M. & Mekalanos, J.J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259, 686–688 (1993).
Walter, J. et al. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 69, 2044–2051 (2003).
Hensel, M. et al. Simultaneous identification of bacterial virulence genes by negative selection. Science 269, 400–403 (1995).
Lee, Y.K., Ho, P.S., Low, C.S., Arvilommi, H. & Salminen, S. Permanent colonization by Lactobacillus casei is hindered by the low rate of cell division in mouse gut. Appl. Environ. Microbiol. 70, 670–674 (2004).
Acknowledgements
Supported in part by a grant from the National Institutes of Health (DK30292).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Sonnenburg, J., Angenent, L. & Gordon, J. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine?. Nat Immunol 5, 569–573 (2004). https://doi.org/10.1038/ni1079
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni1079
This article is cited by
-
The appendix and ulcerative colitis — an unsolved connection
Nature Reviews Gastroenterology & Hepatology (2023)
-
Digestive exophagy of biofilms by intestinal amoeba and its impact on stress tolerance and cytotoxicity
npj Biofilms and Microbiomes (2023)
-
Issues for patchy tissues: defining roles for gut-associated lymphoid tissue in neurodevelopment and disease
Journal of Neural Transmission (2023)
-
Bacteroides rhinocerotis sp. nov., isolated from the fresh feces of rhinoceros in Beijing Zoo
Archives of Microbiology (2023)
-
A mucin-responsive hybrid two-component system controls Bacteroides thetaiotaomicron colonization and gut homeostasis
Journal of Microbiology (2022)