Abstract
Despite a century of research on complex traits in humans, the relative importance and specific nature of the influences of genes and environment on human traits remain controversial. We report a meta-analysis of twin correlations and reported variance components for 17,804 traits from 2,748 publications including 14,558,903 partly dependent twin pairs, virtually all published twin studies of complex traits. Estimates of heritability cluster strongly within functional domains, and across all traits the reported heritability is 49%. For a majority (69%) of traits, the observed twin correlations are consistent with a simple and parsimonious model where twin resemblance is solely due to additive genetic variation. The data are inconsistent with substantial influences from shared environment or non-additive genetic variation. This study provides the most comprehensive analysis of the causes of individual differences in human traits thus far and will guide future gene-mapping efforts. All the results can be visualized using the MaTCH webtool.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Moore, J.H. Analysis of gene-gene interactions. Curr. Protoc. Hum. Genet. Chapter 1, Unit 1.14 (2004).
Hill, W.G., Goddard, M.E. & Visscher, P.M. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4, e1000008 (2008).
Traynor, B.J. & Singleton, A.B. Nature versus nurture: death of a dogma, and the road ahead. Neuron 68, 196–200 (2010).
Zuk, O., Hechter, E., Sunyaev, S.R. & Lander, E.S. The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. USA 109, 1193–1198 (2012).
Phillips, P.C. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9, 855–867 (2008).
Visscher, P.M., Brown, M.A., McCarthy, M.I. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012).
Manolio, T.A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Stranger, B.E., Stahl, E.A. & Raj, T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics 187, 367–383 (2011).
Maher, B. Personal genomes: the case of the missing heritability. Nature 456, 18–21 (2008).
Eichler, E.E. et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11, 446–450 (2010).
Nelson, R.M., Pettersson, M.E. & Carlborg, Ö. A century after Fisher: time for a new paradigm in quantitative genetics. Trends Genet. 29, 669–676 (2013).
Barker, J.S. Inter-locus interactions: a review of experimental evidence. Theor. Popul. Biol. 16, 323–346 (1979).
Cockerham, C.C. An extension of the concept of partitioning hereditary variance for analysis of covariances among relatives when epistasis is present. Genetics 39, 859–882 (1954).
Cockerham, C.C. in Statistical Genetics and Plant Breeding 53–94 (Nat. Acad. Sci. Nat. Res. Council Publ., 1963).
Kempthorne, O. On the covariances between relatives under selfing with general epistacy. Proc. R. Soc. Lond. B Biol. Sci. 145, 100–108 (1956).
Crow, J.F. & Kimura, M. An Introduction To Population Genetics Theory (Harper and Row, 1970).
Carlborg, O. & Haley, C.S. Epistasis: too often neglected in complex trait studies? Nat. Rev. Genet. 5, 618–625 (2004).
Falconer, D.S. & Mackay, T.F.C. Quantitative Genetics (Longman Group, 1996).
Lynch, M. & Walsch, B. Genetics and Analysis of Quantitative Traits (Sinauer Associates, 1998).
Begg, C.B. & Mazumdar, M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101 (1994).
Egger, M., Davey Smith, G., Schneider, M. & Minder, C. Bias in meta-analysis detected by a simple, graphical test. Br. Med. J. 315, 629–634 (1997).
Rosenthal, R. The file drawer problem and tolerance for null results. Psychol. Bull. 86, 91–106 (1979).
Schulze, R. Meta-Analysis: A Comparison Of Approaches (Hogrefe & Huber, 2004).
Drasgow, F. in Encyclopedia of Statistical Sciences (eds. Kotz, S., Read, C.B., Balakrishnan, N. & Vidakovic, B.) Vol. 7, 68–74 (John Wiley & Sons, 2006).
Olsson, U. Maximum likelihood estimation of the polychoric correlation coefficient. Psychometrika 44, 443–460 (1979).
Falconer, D.S. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann. Hum. Genet. 29, 51–76 (1965).
Smith, C. Concordance in twins: methods and interpretation. Am. J. Hum. Genet. 26, 454–466 (1974).
Jiang, H. & Doerge, R.W. Estimating the proportion of true null hypotheses for multiple comparisons. Cancer Inform. 6, 25–32 (2008).
Storey, J.D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445 (2003).
Bulik-Sullivan, B.K. & Sullivan, P.F. The authorship network of genome-wide association studies. Nat. Genet. 44, 113 (2012).
Blondel, V., Guillaume, J., Lambiotte, R. & Lefebvre, E. Fast unfolding of community hierarchies in large networks. J. Stat. Mech. 10, P10008 (2008).
Acknowledgements
We would like to thank M. Frantsen, M.P. Roeling, R. Lee and D.M. DeCristo for their contribution to collecting the full texts of selected twin studies and data entry. This work was funded by the Netherlands Organization for Scientific Research (NWO VICI 453-14-005, NWO Complexity 645-000-003), by the Australian Research Council (DP130102666) and by the Australian National Health and Medical Research Council (APP613601).
Author information
Authors and Affiliations
Contributions
D.P., B.B., P.F.S. and P.M.V. performed the analyses. D.P. conceived the study. D.P., T.J.C.P. and P.M.V. designed the study. T.J.C.P. and D.P. collected and entered the data. D.P. and P.F.S. categorized traits according to standard classifications. A.v.B. and C.A.d.L. checked data entries, and checked and wrote statistical scripts. A.v.B. designed and programmed the webtool. D.P., T.J.C.P. and P.M.V. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Authorship co-occurrence matrix on 2,748 twin studies published between 1958 and 2012.
Each colored cell represents two authors who appeared on the same paper; darker cells indicate authors that co-published more frequently. The filter of at least 25 papers per author was set for readability. The web application MaTCH has an interactive version of this matrix.
Supplementary Figure 2 Funnel plots across all traits for twin correlations and variance components.
Z, Z-converted correlation; MZ, monozygotic twins; DZ, dizygotic twins; DZSS, DZ same-sex twins; MZM, MZ male twins; MZF, MZ female twins; DZM, DZ male twins; DZF, DZ female twins; DOS, DZ opposite-sex twins; h2, heritability; c2, shared environment; h2 same sex; c2 same sex; h2 males; c2 males; h2 female; c2 females.
Supplementary Figure 3 Funnel plots for rMZ across the major trait domains.
The plots denote the relationship between the Z-transformed rMZ and its standard error. SE, standard error.
Supplementary Figure 4 Funnel plots for rDZ across the major trait domains.
The plots denote the relationship between the Z-transformed rDZ and its standard error. SE, standard error.
Supplementary Figure 5 Funnel plots for h2 across the major trait domains.
The plots denote the relationship between the Z-transformed h2 and its standard error. SE, standard error.
Supplementary Figure 6 Funnel plots for c2 across the major trait domains.
The plots denote the relationship between the Z-transformed c2 and its standard error. SE, standard error.
Supplementary Figure 7 Distribution of twin correlations and variance components in full and best models across all traits from 2,748 studies.
rMZ, monozygotic twin correlation; rDZ, dizygotic twin correlation; rDZSS, DZ same-sex twin correlation; rMZM, MZ male twin correlation; rMZF, MZ female twin correlation; rDZM, DZ male twin correlation; rDZF, DZ female twin correlation; rDOS, DZ opposite-sex twin correlation; h2, heritability; c2, shared environment; h2 same sex;c2 same sex; h2 males; c2 males; h2 females; c2 females. “BEST” denotes estimates from the most parsimonious models per study. All other estimates are from “FULL” models.
Supplementary Figure 8 Distribution of differences between MZ and DZ correlations.
rMZ, monozygotic twin correlation; rDZ, dizygotic twin correlation; rDZSS, DZ same-sex twin correlation; rMZM, MZ male twin correlation; rMZF, MZ female twin correlation; rDZM, DZ male twin correlation; rDZF, DZ female twin correlation; rDOS, DZ opposite-sex twin correlation.
Supplementary Figure 11 Scatterplots of all MZ versus DZ correlations.
Contour lines indicate the density of the data in that region. The lines are ‘heat’ colored from blue to red, indicating increasing data density.
Supplementary Figure 12 QQ plots of the χ2 test statistics for testing the null hypothesis that 2(rMZ – rDZ) = 0 and 2rDZ – rMZ = 0 and relationship with sample size.
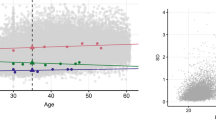
(a) The deviation from the null hypotheses is quantified with the inflation λ in the QQ plots. (b) Effects as a function of sample size.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Note and Supplementary Tables 1–19, 22–24 and 26–31. (PDF 4786 kb)
Supplementary Tables 20, 21, 25, 32 and 33.
Supplementary Tables 20, 21, 25, 32 and 33. (XLSX 609 kb)
Rights and permissions
About this article
Cite this article
Polderman, T., Benyamin, B., de Leeuw, C. et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47, 702–709 (2015). https://doi.org/10.1038/ng.3285
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3285
This article is cited by
-
The genetic architecture of youth anxiety: a study protocol
BMC Psychiatry (2024)
-
Attention-deficit/hyperactivity disorder
Nature Reviews Disease Primers (2024)
-
Genetic effects on variability in visual aesthetic evaluations are partially shared across visual domains
Communications Biology (2024)
-
Examining intergenerational risk factors for conduct problems using polygenic scores in the Norwegian Mother, Father and Child Cohort Study
Molecular Psychiatry (2024)
-
Can flow proneness be protective against mental and cardiovascular health problems? A genetically informed prospective cohort study
Translational Psychiatry (2024)