Abstract
We report sequencing-based whole-genome association analyses to evaluate the impact of rare and founder variants on stature in 6,307 individuals on the island of Sardinia. We identify two variants with large effects. One variant, which introduces a stop codon in the GHR gene, is relatively frequent in Sardinia (0.87% versus <0.01% elsewhere) and in the homozygous state causes Laron syndrome involving short stature. We find that this variant reduces height in heterozygotes by an average of 4.2 cm (−0.64 s.d.). The other variant, in the imprinted KCNQ1 gene (minor allele frequency (MAF) = 7.7% in Sardinia versus <1% elsewhere) reduces height by an average of 1.83 cm (−0.31 s.d.) when maternally inherited. Additionally, polygenic scores indicate that known height-decreasing alleles are at systematically higher frequencies in Sardinians than would be expected by genetic drift. The findings are consistent with selection for shorter stature in Sardinia and a suggestive human example of the proposed 'island effect' reducing the size of large mammals.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Pilia, G. et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2, e132 (2006).
Silventoinen, K., Kaprio, J., Lahelma, E. & Koskenvuo, M. Relative effect of genetic and environmental factors on body height: differences across birth cohorts among Finnish men and women. Am. J. Public Health 90, 627–630 (2000).
Lango Allen, H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 (2010).
Wood, A.R. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014).
Francalacci, P. et al. Low-pass DNA sequencing of 1200 Sardinians reconstructs European Y-chromosome phylogeny. Science 341, 565–569 (2013).
Sidore, C. et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat. Genet. doi: 10.1038/ng.3368 (14 September 2015).
Arcaleni, E. Secular trend and regional differences in the stature of Italians. J. Anthropol. Sci. 90, 233–237 (2012).
Laron, Z. Laron Syndrome—From Man to Mouse (Springer-Verlag, 2011).
Laron, Z. The syndrome of familial dwarfism and high plasma immunoreactive human growth hormone. Birth Defects Orig. Artic. Ser. 10, 231–238 (1974).
Rosenbloom, A.L., Guevara Aguirre, J., Rosenfeld, R.G. & Fielder, P.J. The little women of Loja—growth hormone–receptor deficiency in an inbred population of southern Ecuador. N. Engl. J. Med. 323, 1367–1374 (1990).
Laron, Z., Klinger, B., Erster, B. & Silbergeld, A. Serum GH binding protein activities identifies the heterozygous carriers for Laron type dwarfism. Acta Endocrinol. (Copenh.) 121, 603–608 (1989).
Guevara-Aguirre, J. et al. Effects of heterozygosity for the E180 splice mutation causing growth hormone receptor deficiency in Ecuador on IGF-I, IGFBP-3, and stature. Growth Horm. IGF Res. 17, 261–264 (2007).
Lanktree, M.B. et al. Meta-analysis of dense genecentric association studies reveals common and uncommon variants associated with height. Am. J. Hum. Genet. 88, 6–18 (2011).
Lee, M.P. et al. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc. Natl. Acad. Sci. USA 96, 5203–5208 (1999).
Soejima, H. & Higashimoto, K. Epigenetic and genetic alterations of the imprinting disorder Beckwith-Wiedemann syndrome and related disorders. J. Hum. Genet. 58, 402–409 (2013).
Horikoshi, M. et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat. Genet. 45, 76–82 (2013).
Johnson, A.D. et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat. Genet. 42, 608–613 (2010).
Newton-Cheh, C. et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat. Genet. 41, 399–406 (2009).
Voight, B.F. et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 42, 579–589 (2010).
Kong, A. et al. Parental origin of sequence variants associated with complex diseases. Nature 462, 868–874 (2009).
Orrù, V. et al. Genetic variants regulating immune cell levels in health and disease. Cell 155, 242–256 (2013).
Turchin, M.C. et al. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat. Genet. 44, 1015–1019 (2012).
Berg, J.J. & Coop, G. A population genetic signal of polygenic adaptation. PLoS Genet. 10, e1004412 (2014).
Steinthorsdottir, V. et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat. Genet. 46, 294–298 (2014).
Danjou, F. et al. Genome-wide association analyses based on whole-genome sequencing in Sardinia provide insights into regulation of hemoglobin levels. Nat. Genet. doi: 10.1038/ng.3307 (14 September 2015).
Millien, V. Morphological evolution is accelerated among island mammals. PLoS Biol. 4, e321 (2006).
van der Geer, A., Lyras, G., de Vos, J. & Dermitzakis, M. in Evolution of Island Mammals 103–130 (Wiley-Blackwell, 2010).
Mathieson, I. et al. Eight thousand years of natural selection in Europe. bioRxiv 016477 (2015).
Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014).
Haak, W. et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015).
Pistis, G. et al. Rare variant genotype imputation with thousands of study-specific whole-genome sequences: implications for cost-effective study designs. Eur. J. Hum. Genet. 23, 975–983 (2015).
Kang, H.M. et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354 (2010).
Szpiech, Z.A. & Hernandez, R.D. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 31, 2824–2827 (2014).
Abecasis, G.R., Cherny, S.S., Cookson, W.O. & Cardon, L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
McVicker, G., Gordon, D., Davis, C. & Green, P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 5, e1000471 (2009).
Acknowledgements
We thank all the volunteers who generously participated in this study and made this research possible. All participants provided informed consent, and the studies were approved by local research ethic committees: Comitato Etico di Azienda Sanitaria Locale 8, Lanusei (2009/0016600) and Comitato Etico di Azienda Sanitaria Locale 1, Sassari (2171/CE). This study was funded in part by the US National Institutes of Health (National Institute on Aging, National Heart, Lung, and Blood Institute, and National Human Genome Research Institute). This research was supported by National Human Genome Research Institute grants HG005581, HG005552, HG006513, HG007089, HG007022 and HG007089; by National Heart, Lung, and Blood Institute grant HL117626; by the Intramural Research Program of the US National Institutes of Health, National Institute on Aging, contracts N01-AG-1-2109 and HHSN271201100005C; by Sardinian Autonomous Region (L.R. 7/2009) grant cRP3-154; by grant FaReBio2011 'Farmaci e Reti Biotecnologiche di Qualità'; by the PB05 InterOmics MIUR Flagship Project; by a US National Institutes of Health National Research Service Award (NRSA) postdoctoral fellowship (F32GM106656) to C.W.K.C.; by UC MEXUS-CONACYT doctoral fellowship 213627 to D.O.D.V.; and by Italian Ministry of Education, University and Research (MIUR) grant 5571/DSPAR/2002. The HELIC study was funded by the Wellcome Trust (098051) and the European Research Council (ERC-2011-StG 280559-SEPI). The TEENAGE study has been supported by the Wellcome Trust (098051), European Union (European Social Fund (ESF)) and Greek national funds through the Operational Programme 'Education and Lifelong Learning' of the National Strategic Reference Framework (NSRF) research funding programme Heracleitus II, Investing in Knowledge Society Through the European Social Fund. The UK Household Longitudinal Study is led by the Institute for Social and Economic Research at the University of Essex and funded by the Economic and Social Research Council. Information on how to access the data can be found on the Understanding Society website (https://www.understandingsociety.ac.uk/). This study makes use of data generated by the UK10K Consortium, derived from samples from UK10K_COHORTS_TWINSUK (the TwinsUK cohort) and UK10K_COHORT_ALSPAC (the Avon Longitudinal Study of Parents and Children cohort). A full list of the investigators who contributed to the generation of the data is available from http://www.UK10K.org/. Funding for UK10K was provided by the Wellcome Trust under award WT091310. We thank J. Berg for scripts and suggestions on the polygenic score analysis.
Author information
Authors and Affiliations
Consortia
Contributions
M.Z., G.R.A., J.N., D.S. and F.C. conceived and supervised the study. M.Z., C.S., C.W.K.C., J.N., D.S. and F.C. drafted the manuscript. S.S., K.E.L. and G.R.A. revised the manuscript and wrote specific sections of it. A.A., C.J. and R.L. supervised sequencing experiments. F.B. and A. Maschio performed sequencing experiments. C.S., M.S., M.M. and S.S. carried out genetic association analyses. C.S. analyzed DNA sequence data. M.Z., A. Mulas, F.B., S.U. and R.N. carried out SNP array genotyping. M.Z. and A. Mulas verified genotypes by TaqMan genotyping. J.H.M., C.W.K.C., M.S., M.F., D.O.D.V., K.E.L. and J.N. performed polygenic score and related population genetic analyses. A. Meloni and A.D. performed clinical characterization of Laron carriers. S.V. provided DNA for the Sardinian replication sample set. F.M., M.P.C., G.B., M.S. and S.S. performed replication analysis. N.S., N.J.T., G.D., I.T., E.Z. and the UK10K group provided KCNQ1 fine-mapping data. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of members and affiliations appears in the Supplementary Note.
A full list of members and affiliations appears in the Supplementary Note.
Integrated supplementary information
Supplementary Figure 1 GWAS on height for SardiNIA cohort.
(a) Schematic of the sequencing-based GWAS approach. (b) Quantile-quantile plots for the ~12 million variants tested. The black line shows the expected P values under the null distribution. Colored lines represent observed P values for different ranges of frequency. Green circles, MAF = 0.05–0.5; red triangles, MAF = 0.01–0.05; blue squares, MAF = 0.005–0.01; gray dots, MAF = 0.001–0.005. (c) Manhattan plot of the association P values. The y axis shows the association strength (–log10 (P value)) versus the genomic position on the x axis.
Supplementary Figure 2 Height distribution within each genotype class for the top associated variants at GHR and KCNQ1.
(a) The height residual distribution within each genotype class after normalization for age and sex for rs121909358 (chr5:42689036). (b) The height residual distribution within each genotype class for each parent of origin after normalization for age and sex for rs150199504 (chr11:2814960).
Supplementary Figure 3 Haplotype blocks in the region surrounding the Laron variant (rs121909358).
The figure shows the extent of haplotype similarity for the 11 unrelated sequenced carriers. Each of the two chromosomes is plotted separately in the upper panel (if carrying the reference/ancestral allele) and lower panel (if carrying the alternate/derivate allele). Colors represent identical haplotypes; the extension of the haplotype around the core segment bearing the Laron variant varies because of recombination events. The x axis shows the distance from rs12190358 (chr5:42689036).
Supplementary Figure 4 Polygenic score analysis for height.
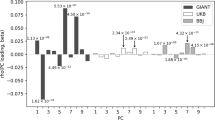
(a) The analysis of polygenic score using effect sizes published by the GIANT Consortium. (b) The polygenic score analysis using 162 SNPs immune to population stratification22 and in LD (r2 > 0.5) with the height loci published by the GIANT Consortium.
Supplementary Figure 5 Average frequency difference for height-decreasing alleles between European populations.
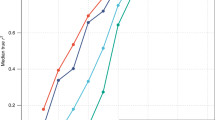
The figure shows the average frequency difference between the 1000 Genomes Project CEU population (taken as the reference), the SardiNIA cohort (SDI) and other 1000 Genomes Project populations (IBS, TSI, GBR and FIN). The frequency difference is evaluated for the height-decreasing allele of the 691 variants reported in the GIANT Consortium meta-analysis4.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1–4 and Supplementary Note. (PDF 1805 kb)
Source data
Rights and permissions
About this article
Cite this article
Zoledziewska, M., Sidore, C., Chiang, C. et al. Height-reducing variants and selection for short stature in Sardinia. Nat Genet 47, 1352–1356 (2015). https://doi.org/10.1038/ng.3403
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3403
This article is cited by
-
MTHFR, XRCC1 and OGG1 genetic polymorphisms in breast cancer: a case-control study in a population from North Sardinia
BMC Cancer (2020)
-
Searching for parent-of-origin effects on cardiometabolic traits in imprinted genomic regions
European Journal of Human Genetics (2020)
-
Parent-of-origin effects on quantitative phenotypes in a large Hutterite pedigree
Communications Biology (2019)
-
Complex Phenotypes: Mechanisms Underlying Variation in Human Stature
Current Osteoporosis Reports (2019)
-
Characterizing rare and low-frequency height-associated variants in the Japanese population
Nature Communications (2019)