Abstract
Methods to detect polygenic adaptation have recently been shown to be sensitive to uncorrected stratification in GWAS, thereby casting doubts on whether polygenic adaptation is prevalent among humans. Consistent with a signal of adaptation at human height loci, the mean FST among African, East Asian, and European populations was shown to be significantly higher at height-associated SNPs than that at non-associated SNPs. This conclusion was reached, however, using height-associated SNPs ascertained from a GWAS design impacted by residual confounding due to uncorrected stratification. Specifically, we show here that the estimated effect sizes are significantly correlated with population structure across continents, potentially explaining the elevated differentiation previously reported. We alleviated these concerns of confounding by ascertaining height-associated SNPs from two biobank GWAS (UK Biobank, UKB, and Biobank Japan, BBJ), where measures to control for confounding in GWAS are more effective. Consistent with a global signature of polygenic adaptation, we found that compared to non-associated SNPs, frequencies of height-associated SNPs are indeed significantly more differentiated among continental populations from both the 1000 Genomes Project (p = 0.0012 for UKB and p = 0.0265 for BBJ), and the Human Genome Diversity Project (p = 0.0225 for UKB and p = 0.0032 for BBJ). However, we found no significant difference among continental populations in polygenic height scores. Through simulations, we found that polygenic score-based statistics could lose power in detecting polygenic adaptation in presence of independent converging selections, thereby potentially explaining the inconsistent results based on FST and polygenic scores.
Similar content being viewed by others
Introduction
Given the highly polygenic nature of many human complex traits, polygenic adaptation was thought to be an important mechanism of phenotypic evolution in humans. Under this model, only a subtle but coordinated allelic or haplotypic signature across the causal loci underlying the selected trait is expected [1]. In humans, one of the earliest putative examples of polygenic adaptation was based on an inferred signal of adaptation at height-associated SNPs [2,3,4,5,6]. Though height itself may not be the target of selection, these early reports suggested that natural selection contributed to the differentiation of height between human populations.
While most of these studies evaluated the adaptive signature between populations from the European continent, one study suggested polygenic adaptation as one of the reasons for differences in human height among global populations by evaluating allele frequency differentiations at height-associated SNPs [6]. Specifically, Guo et al. [6] demonstrated that compared to randomly selected, frequency- and LD-score-matched SNPs, height-associated SNPs showed significantly higher mean FST across the three continental populations (Africans, Europeans, and East Asians). However, height-associated SNPs examined in Guo et al. were ascertained from a genome-wide association studies (GWAS) performed by the Genetic Investigation of Anthropometric Traits (GIANT) consortium [7]. It has recently been suggested that because of residual uncorrected stratification, the estimated effect sizes of SNPs detected in GIANT showed a subtle but biased correlation with population structure in Europe. As a result, polygenic scores (PSs) constructed on the basis of these SNPs showed exaggerated difference in human height between Northern and Southern Europeans [8,9,10]. Therefore, there is concern whether the previous polygenic signals among global populations were confounded by population stratification, and it remains an open question whether polygenic adaptative signals at height-associated SNPs are observed in any human populations beyond a few populations with special population history [10, 11].
We aimed to determine if frequencies at height-associated SNPs are indeed more differentiated among continental populations. If true, our results would lend support to adaptation at a global scale. On the one hand, there are reasons to believe that conclusions from Guo et al. may be robust to residual stratification in GWAS since FST, the main statistics used by Guo et al. to measure allele frequency differentiations, is unsigned and does not rely on the estimated effect sizes. On the other hand, while population stratification within Europe is not expected to bias allele frequency differentiations at height-associated SNPs among global populations, it is possible that structures among global populations are correlated with structures within Europe due to, for example, migration and admixture between Europeans and populations outside of Europe. This could result in spurious signals of adaptation.
In the present study, we used summary statistics from UKB and BBJ to re-examine whether height-associated SNPs exhibit signs of adaptation among the three continental populations (Africans, Europeans, and East Asians). We demonstrated that the effect sizes of height-associated SNPs ascertained from UKB and BBJ showed little or no association with population structure across the three continents, alleviating any concerns of confounding due to residual stratification. Using this approach, we found that allele frequencies of height-associated SNPs remain significantly differentiated among continental populations when compared to that of non-associated SNPs, consistent with a polygenic adaptive signal at these SNPs [6]. However, applying two PS-based testing frameworks [3, 4] to test for polygenic adaptation, we detected no significant difference in height PSs among the three populations and observed that even the ranked order of PSs among populations appeared sensitive to the choice of variants for analysis. PSs as constructed based on our knowledge from existing GWAS predict any given complex trait poorly both within or between populations [12, 13], which could lead to a loss of power in PS-based framework. We examined this possibility through simulation, but found that PS-based methods maintain higher power to detect polygenic adaptation across a range of different strength of selection. Instead, through simulations we showed that PS-based framework could lose power if different parts of the trait architecture is under selection in each of the populations undergoing independent convergent evolution. In this scenario FST-based statistic would be more powered to detect evidence of adaptation.
Materials and methods
GWAS panels and population genetic datasets
We obtained publicly available GWAS summary statistics from three studies: [1] GIANT consortium [2], UK Biobank (UKB), and [3] Biobank Japan (BBJ). Details of each GWAS dataset can be found in Supplementary Material. To evaluate polygenic selection at height-associated SNPs across continental populations, we analyzed samples from Africa, East Asia, and Europe from 1000 Genomes phase 3 release [14], including five African subpopulations, four European subpopulations, and five East Asian subpopulations. We did not include the two admixed African subpopulations (ACB and ASW; see Fig. S1) and the Finnish European subpopulation (FIN) because of its known unique demographic history [15, 16]. We also replicated our analysis on the Human Genome Diversity Project (HGDP) dataset [17], using either all 929 samples from seven regions (including 104 Africans, 61 Americans, 197 Central South Asians, 223 East Asians, 155 Europeans, 161 Middle East, and 28 Oceanians), or 482 samples from three continents (104 Africans, 223 East Asians, and 155 Europeans). Details of populations from 1000 Genomes and HGDP can be found in Tables S1, S2.
Population structure analysis
PCAs were performed by applying Eigenstrat (version 7.2.1) on a dataset of biallelic SNPs with MAF > 5% in the dataset, thinned to no more than one SNP in 200 kb, and removed SNPs in known regions of long-range LD [18]. We measured potential confounding due to residual stratification as the correlation between estimated SNP effect sizes from GWAS summary statistics with its loading on a particular PC, as previously described [8, 10]. A strongly significant correlation implies a systematic bias in effect sizes that aligns with the structure captured by a particular PC, which could in turn confound the inference of polygenic adaptation. We assessed the significance of the correlation on the basis of jackknife standard errors computed by splitting the genome into 1000 blocks with an equal number of variants.
Signature of selection at height-associated SNPs
To ascertain height-associated variants, we selected a set of genome-wide significant variants (p < 5e−8) with MAF > 1% in GWAS panel after greedily pruning any other variants such that no two variants were within 1 Mb of each other. We then further pruned by LD using 1000 Genomes as the reference such that no two variants would have a r2 > 0.1. We used CEU, GBR, and JPT data as reference LD for pruning GIANT, UKB, and BBJ summary statistics, respectively. In total, we identified 458, 709, and 407 independent height-associated SNPs from GIANT, UKB, and BBJ summary statistics (Table S3 and Fig. S2). To evaluate the evidence of selection at height-associated SNPs, we applied the following two approaches: FST enrichment test and PS-based tests.
FST enrichment test
Wright’s fixation index (FST) was calculated from PLINK [19]. Following Guo et al. [6] we compared the mean FST value of the height-associated SNPs with that of the non-associated SNPs (i.e., p > 5e−8), which were presumed to be neutral, randomly selected from the rest of the genome matched by MAF and LD score. To randomly identify matched SNPs, for the summary statistics from each GWAS panel we divided all SNPs into 25 bins based on MAF in increments of 0.02, but excluding SNPs with MAF < 0.01. Within each MAF bin, we further divided SNPs into 20 bins of LD scores in increments of 5% quantiles, for a total of 500 bins. LD scores were computed as the sum of the LD r2 between the focal SNP and all the flanking SNPs within 10-Mb window. We used 1000 Genomes EUR (excluding FIN) as reference LD for GIANT and UKB, and used EAS for BBJ. For each height-associated SNP, we then randomly draw one matched SNP from the same bin. Ten thousand sets of matched SNPs were drawn to generate a distribution of mean FST under presumed neutrality to derive a two-tailed empirical p value.
PS-based analysis
We estimated the PS for each population as the sum of allele frequencies at a set of L height-associated SNPs (L = 458, 709, and 407 for GIANT, UKB, and BBJ, respectively) weighted by effect sizes from each GWAS panel (i.e., \(Z \,=\, \mathop {\sum}\nolimits_{l \,=\, 1}^L {2\beta _lp_l}\), where pl and βl were the allele frequency and effect size at SNP l). We conducted the QX test to determine whether the estimated PSs exhibited more variance among populations than null expectation under genetic drift, and computed the conditional Z score to identify outlier populations and regions which contributed to the excess of variance; both statistics were previously described [3]. The scripts were downloaded from GitHub repository of the original authors (see https://github.com/jjberg2/PolygenicAdaptationCode).
We also adopted the PS-based method used in Guo et al. [6] which was originally proposed by Robinson et al. [4] to estimate the deviation of the PS of a population from the overall mean. We calculated the PS for each individual as \(g \,=\, \mathop {\sum}\nolimits_{l \,=\, 1}^L {\beta _lx_l}\), where x represents the count of alleles with respect to the effect size of each SNP (0, 1, or 2). We then standardized g on the scale of all test populations, and computed its mean value for each population. The deviation of the observed mean PS from neutrality was assessed empirically by comparing to the distribution under presumed neutrality based on 10,000 sets of randomly sampled matched SNP as described in the FST enrichment analysis.
Forward simulation
We performed forward simulations using SLiM version 3 [20] to investigate two potential reasons for the apparent inconsistent conclusions of adaptation using the FST-based and PS-based methods: poor prediction of phenotype among ascertained trait-associated SNPs and convergent evolution in multiple populations. We assumed a model of three constant-sized populations (Fig. S3), and a genetic architecture of complex trait dictated by 120 independent loci shared across all three populations (P1, P2, and P3). Each locus is 100 kb in length, and harbors exactly one causal variant. In the first simulated scenario, all 120 SNPs were selected in one population (P1). To test the impact due to poor prediction accuracy of PS, for each of the 120 selected loci we hid the causal SNP and randomly sampled another SNP in the same locus as the ascertained trait-associated SNP. This represented an extreme case of ascertainment where the GWAS may inform the correct locus, but was uninformative of the actual causal allele. Expectedly, this drastically reduced the prediction accuracy of the resulting PS. We then compared the power of FST and QX using these ascertained SNPs. To test the impact due to convergent evolution, we further simulated scenarios of adaptation where subsets of the 120 SNPs were selected in two (P1 and P2) or all three populations. We then evaluated the power of detecting selection by applying the FST or QX framework to the set of selected causal SNPs. In this case, we effectively removed any concerns of poor prediction accuracy due to PS. In all simulated scenarios, we evaluated the power of FST and QX at a p value threshold of 0.05 across a range of selective coefficient from 1e−2 to 1e−4. Details of the demographic model and simulation parameters can be found in Supplementary Material.
Results
Population stratification
It has been previously shown that the set of height-associated SNPs analyzed by Guo et al. were drawn from genome-wide summary statistics where the effect sizes significantly correlated with population structure within Europe [8,9,10]. If the effect sizes estimated from the GWAS study genome-wide are also significantly correlated with global population structure, then the conclusions from Guo et al. could potentially be spurious. We thus first evaluated the impact of global population stratification on effect sizes estimated from different GWAS panels: the GIANT consortium, the UKB, and the BBJ datasets. We conducted PCAs across continents (Africa, Europe, and East Asia, Fig. S4), within Europe (Fig. S5), and within East Asia (Fig. S6) using data from 1000 Genomes, and examined the correlation between effect sizes (of all SNPs with MAF > 1%, regardless of p values for association with height) and the PC loading on those PCAs. We recapitulated previous findings [10] that GIANT effect sizes were significantly correlated with the loading of the first two PCs within Europe (Fig. S7 and Table S4). We also observed that BBJ effect sizes showed a small but significant correlation with the first two PCs in the analysis within East Asia (Fig. S8 and Table S4), suggesting that measures to control for population structure in BBJ were not completely effective in controlling for stratification within the East Asian continent, although the effect sizes estimated in BBJ are uncorrelated with population structure within Europe presumably due to its geographic distance [10].
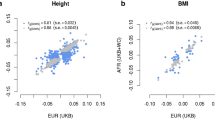
In the analysis across continents, the first two PCs reflected respectively the differentiation between Africans and Eurasians and between Europeans and East Asians (Fig. S4). We found that the effect sizes estimated in GIANT were highly correlated with the loading of the first two PCs of population structure (rho = 0.026, p = 1.13e−8 for PC1; rho = −0.086, p = 1.62e−78 for PC2), as well as with lower PCs that are driven by within-European structure (PC6 and PC8) or within-African structure (PC5) (Figs. 1, S9, S4, and Table S4). This suggests that even though consortium GWAS were conducted in European-ancestry populations, residual stratification can lead to a correlation between effect sizes of SNPs with inter-continental structures. Because FST measures are also strongly associated with PC loadings (Table S5), the elevated differentiation among height-associated SNPs as reported by Guo et al. could potentially be a confounded observation.
Restricting to all SNPs with MAF > 1% (regardless of p values for association with height) in each GWAS panel, Pearson correlation coefficients of PC loadings and SNP effects from GIANT, UKB, and BBJ were computed. Twenty PCs were computed in Africans, Europeans, and East Asians from 1000 Genomes; only the first 10 are shown here for readability, see Fig. S9 and Table S4 for more PCs. P values are based on jackknife standard errors (1000 blocks). P values lower than 0.05/20 are indicated on each bar.
Compared to the strong correlations between effect sizes of SNPs from GIANT with inter-continental structures, the correlations were much smaller and insignificant in UKB (rho = 0.0016, p = 0.683 for PC1; rho = −0.0051, p = 0.107 for PC2) and BBJ (rho = −7.15e−4, p = 0.831 for PC1; rho = 0.0013, p = 0.673 for PC2) (Fig. 1), suggesting that UKB and BBJ summary statistics are not likely to be affected by population stratifications across continents. Even when there were significant associations between effect sizes and PC loadings of lower PCs, the magnitudes of the associations were much smaller (Fig. 1). The fact that those lower PCs were driven by within-continent structures that were independent from inter-continent structures (Fig. S4) suggests that population stratification effect caused by those lower PCs is not expected to bias the signals of polygenic adaptation at the cross-continental scale. Therefore, we conclude that height-associated SNPs ascertained from UKB and BBJ can be used to test the robustness of conclusions from Guo et al. based on FST differentiation.
Enrichment of FST in height-associated SNPs
On the basis of independent SNPs associated with height with p < 5e−8 in UKB (709 SNPs) and BBJ (407 SNPs), we robustly tested whether height-associated SNPs were more differentiated across continental populations than matched non-associated SNPs, as measured by FST. We found that the mean FST values for the height-associated SNPs ascertained from UKB and BBJ remained significantly higher than those of the matched, presumed neutral, SNPs (p = 0.0012 in UKB and p = 0.0265 in BBJ; Fig. 2), consistent with a signal of adaptation at these SNPs. The signature using the height-associated SNPs ascertained from GIANT was weaker (p = 0.0621) compared to that reported previously (p = 4.93e−6) [6]. This is likely due to the previous authors using less stringent pruning parameter, less stringent p value threshold, and including admixed 1000 Genomes populations in the previous analysis. Consequently, our analysis used ~450 height-associated SNPs for analysis, compared to ~1100 SNPs used in analysis in Guo et al. potentially decreasing our power. We did find a stronger signal (Fig. S10) when we increased the p threshold for ascertaining height-associated SNPs to 5e−6 (681 SNPs), the same value was used in Guo et al. [6]. We also replicated our finding using data from HGDP. The mean FST of the height-associated SNPs remained significantly larger than that of matched, non-associated SNPs on the basis of summary statistics from UKB and BBJ (p = 0.0225 and p = 0.0032, respectively, for the analysis on the three continental populations and p = 0.0051 and p = 0.0002, respectively, for the analysis on the seven regional populations; Fig. S11).
PS-based analysis
We used Berg and Coop’s QX and Conditional Z score framework to evaluate the significance of differences in PSs across populations and continents. We observed a clear signal of adaptation when we used height-associated SNPs ascertained from GIANT (p = 3.01e−6 for QX test). At the continental level, Europeans were significantly taller than would be expected under neutral drift given their genetic relationship with Africans and East Asians and the PSs in Africans and East Asians (p = 1.67e−14 for Conditional Z score); and East Asians were significantly shorter than neutral expectation (p = 3.11e−4 for Conditional Z score). However, the signals disappeared when we used height-associated SNPs ascertained from UKB (p = 0.278 for QX test) or BBJ (p = 0.426 for QX test) (Fig. 3), suggesting the signal from QX analysis may be largely driven by uncorrected stratification in the GIANT data. Moreover, the ranked orders of populations based on PSs were also variable across GWAS panels, consistent with previous reports of poor prediction accuracy of PS models across populations [12]. For example, among the three continents, Europeans appeared to have the highest mean PS based on the height-associated SNPs ascertained from UKB, while Africans had the highest mean PS based on the SNPs ascertained from BBJ. Moreover, using height-associated SNPs ascertained from GIANT coupled with effect size estimates from UKB, we found a significant, but much attenuated, signal among continents (Fig. S12), suggesting that the stratification effect in GIANT came from both the biased ascertainment of height-associated SNPs and the biased estimates of effect sizes, consistent with previous observation [21]. Using Robinson et al.’s PS-based approach, we also observed the adaptive signature using height-associated SNPs ascertained from GIANT but not from UKB or BBJ (Fig. S13).
The PSs were constructed on the basis of the height-associated SNPs ascertained from GIANT (A), UKB (B), and BBJ (C) GWAS summary statistics. Pval (QX) denotes the p value for QX test. The p value for conditional Z score is represented by the size of each circle for each population, and two populations with p lower than 0.01 are CEU (p = 9.52e−5) and IBS (p = 0.0056) in the analysis using GIANT summary statistics; the p value is represented by the thickness of each horizontal solid line for each continent, and those lower than 0.01 are shown in the plot. The horizontal dashed line denotes the expected PS for each continent. African populations: ESN, GWD, LWK, MSL, and YRI; East Asian populations: CDX, CHB, CHS, JPT, and KHV; European populations: CEU, GBR, IBS, and TSI.
We repeated the QX and Conditional Z score analysis using height-associated SNPs ascertained from UKB and BBJ in the three continental populations and the seven regional populations from HGDP and found no signature of selection at either level (Figs. S14, S15), recapitulating our observation from 1000 Genomes. In the HGDP analyses, the only consistently discernable signal of selection are observed in Sardinians, confirming our previous result that shorter height among Sardinians compared to other European populations may be a result of natural selection [10]. Taken together, PS-based analysis in this context appears to be susceptible to the choice of SNPs used in the analysis. While we observed a robust signature of differentiation using FST among continental populations, the adaptive signatures based on PS calculated from GIANT-ascertained SNPs disappeared when we used UKB- or BBJ-ascertained SNPs.
Power to detect polygenic adaptation in simulation
To better understand the reason for a seemingly discrepant conclusion on the presence of polygenic adaptation using either PS-based and FST-based tests, we investigated in simulation two possible explanations: poor prediction accuracy of current height PS and convergent evolution in multiple populations. We simulated multiple scenarios of polygenic selection on a quantitative complex trait controlled by 120 loci where we know the complete genetic architecture to disentangle between the two possible explanations (Methods).
Previous studies have shown that QX test have greater power than FST-based test [3]. We recapitulate this finding in our simulated scenario. When only one population is under selection, and when the genetic architecture is known perfectly, we found that across the spectrum of selection strengths the QX test generally attained equal or greater power than the FST test (Fig. 4A).
A One population under selection, (B) two populations under selection, and (C) three populations under selection. In these simulations, the complete genetic architecture is known. Please refer to Fig. S17 for results when the genetic architecture is not completely known and imperfectly ascertained.
We then first investigated whether poor prediction accuracy of PS could cause Qx test to lose power to detect polygenic selection relative to the FST-based method, thereby giving rise to the apparent discrepancy we observed between the two methods. We took our simulated scenario where only one population is under selection, but the trait-associated SNPs were imperfectly ascertained such that the prediction accuracy was low, particularly in the population that was not used for ascertainment (Methods; Fig. S16). In contrast to when the genetic architecture is perfectly ascertained, when inferring the signature of polygenic adaptation on these poorly ascertained SNPs, both FST and QX showed lower power (Compare Fig. S17 to Fig. 4A). However, QX test still generally attained greater power than FST across the spectrum of selective strengths (Fig. S17).
We then evaluated through simulation the impact due to convergent evolution in multiple populations as the reason for discrepant inference between the PS-based and FST-based tests. Here we simulated either two or three populations under selection. In each scenario, a mutually exclusive subset of the 120 trait loci are under selection in each population (Methods). We assumed that the selected causal SNP is known, so that poor prediction accuracy due to PS would not be a factor. We found that when two populations are under selection, QX test also had more power than FST to detect selection across the range of selective coefficients (Fig. 4B), although the conditional Z statistics would often indicate that the wrong population is under selection (Fig. S18). When all three populations are under selection, we observed that across the range of selection coefficient (from s = 5e−3 to 6e−4), FST attained greater power compared to QX (Fig. 4C).
Discussion
By ascertaining height-associated SNPs from the UKB and BBJ GWAS panels, we showed that the effect sizes of these SNPs were not impacted by population structure across three continents (i.e., Africa, Europe, and East Asia, Fig. 1). Using these two ascertained sets of height-associated SNPs, our study showed that height SNPs appear to be more differentiated among the three continental populations, compared to matched SNPs. Results from our FST-based test are qualitatively consistent with the previous study based on summary statistics from GIANT [6] (Figs. 2, S11) and support a global signature of polygenic adaptation at height-associated SNPs. Although the signal of adaptation at height-associated SNPs is robust, because FST is unable to indicate the direction of differentiation between populations, it is still unclear which of the population(s) we examined experienced selection in the past and contributed to the adaptive signal. PS-based methods applied to multiple populations were expected to help address this question. Assuming the PSs are predictive of the phenotype in each population, a follow-up within-population analysis, such as one using trajectory of PSs over time [22] could complement the QX statistics and clarify the population under selective pressure [10]. However, we could not identify significant differentiation in PS among the three continental populations, whether we used the QX test or Robinson et al.’s PS framework (Figs. 3, S13, S14, S15). Therefore, PS-based methods in the context of global populations appear to be particularly susceptible to the choice of SNPs used in the analysis and could not provide a robust signal indicating the populations under selection.
The discrepancy in the inference resulting from the FST-based test and PS-based test could be attributed to at least two possible reasons. The first reason could be the poor prediction accuracy of PS across populations, because of factors such as the divergence of causal allele frequency, differences in LD between populations, low heritability, or underpowered GWAS. While PSs for human complex traits have been shown to be significantly correlated with the observed phenotype within population [12, 23, 24], prediction accuracies of these PS models within or between populations are currently poor [12, 13, 25,26,27]. Poorly predictive PSs could in principle severely impact the power and interpretation of natural selection inferences based on them. Therefore, it may be a good practice for inference of polygenic adaptation based on PS to first demonstrate that the scores used are sufficiently predictive of the phenotype being studied. Although we did not show directly the PS constructed are sufficiently predictive of height among the non-European populations assessed in this study because of the lack of phenotypic information in the 1000 Genomes or HGDP, other studies have shown generally a significant, albeit less efficacious, correlation between PS informed by Euro-centric GWAS and height in non-European populations [25, 27, 28]. Furthermore, in principle, FST-based inference of polygenic adaptation could also be affected by the same factors leading to poor prediction accuracy of PS, so the poor prediction accuracy may not be able to explain the discordant inference of polygenic selection we observed using PS-based vs. FST-based tests. Indeed, as we showed in simulations, PS-based test in general attained greater power than FST-based test despite the poor prediction accuracy (Fig. S17).
The second reason we investigated is the genetic redundancy of polygenic traits. Because a single polygenic trait corresponds to a large number of causal loci, independently adaptive populations could converge to the same trait optimum using different sets of SNPs [29]. As a result, it could lead to a loss of power of PS-based methods to detect polygenic adaptation if more than one population is under selection. To illustrate this potential, in the simulation scenario with independent selections among multiple populations on the same genetic architecture of a complex trait, we found QX statistics could lose power to infer the presence of adaptation while FST remains more robust (Fig. 4C). Although lower statistical power is not necessarily the only explanation for a negative result, our simulations do suggest that genetic redundancy could be a plausible explanation for our observations here for human height, as the trait architecture is likely largely shared among all humans [30,31,32,33] but recent adaptations could occur independently in multiple populations; we know of at least two instances of adaptive signatures at height-associated SNPs, in Sardinia [10, 34] and in Flores [11], with other possible examples [8,9,10, 22].
Recent reports [8,9,10] suggested that residual uncorrected stratification in GWAS summary statistics from the GIANT consortium may have led to the over-estimated signal of polygenic adaptation at height-associated SNPs in the past, even though effect size estimates from GIANT are highly correlated with those from UKB [8, 9] and BBJ [32]. We similarly confirmed that among SNPs with MAF > 1% in both GIANT and one of the biobank datasets, the effect sizes of height-associated SNPs (p < 5e−8) in GIANT are highly correlated with that found in the biobank summary statistics (Pearson’s r = 0.98, p = 1.72e−300 when compared to UKB and r = 0.87, p = 1.55e−126 when compared to BBJ). These reports generally focused on the context of within-Europe analysis. Here we demonstrated that the effect size estimates in the GIANT summary statistics also exhibit a correlation with global structure (Fig. 1). We speculate that this correlation could stem from shared history and continuous gene flow between Europeans and other populations [35, 36]. For example, there is evidence of recent gene flow (~10 generations ago) from North Africa leading to the south-to-north latitudinal gradient of North African ancestry in Europe [37], which may facilitate the correlation in allele frequencies between the European and African continents. Given the interconnectedness among human populations across continents, it seems that the potential of residual stratification in a consortium GWAS to confound analyses across multiple continental populations should not be ignored.
Single-population biobank-level GWAS (such as UKB and BBJ) seemingly could statistically guard against this level of biases from an analysis standpoint, but these datasets are restricted to a single population. For many diverse populations, a large biobank is currently not feasible and their inclusion in large-scale consortium GWAS will be, for the time being, most equitable and beneficial until research infrastructures are established in these populations. The potential for genetic redundancy of a trait under selection as explored here would support greater inclusions in genetic analysis, as geographically diverse populations would provide opportunities to probe into different aspects of the trait architecture. There are multitudes of other reasons to include more diverse populations into GWAS. However, as we learned in the study of polygenic adaptation it is already difficult to control for stratification in a within-continent population such as European-ancestry individuals, and the problem of stratification will only be exacerbated when we examine rarer and rarer variants [38, 39]. Therefore, we should bear in mind the interconnectedness across human populations, carefully characterize our genetic findings, and thread carefully when interpreting signals of selection [40].
References
Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20:R208–15.
Turchin MC, Chiang CW, Palmer CD, Sankararaman S, Reich D, Hirschhorn JN. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat Genet. 2012;44:1015–9.
Berg JJ, Coop G. A population genetic signal of polygenic adaptation. PLoS Genet. 2014;10:e1004412.
Robinson MR, Hemani G, Medina-Gomez C, Mezzavilla M, Esko T, Shakhbazov K, et al. Population genetic differentiation of height and body mass index across Europe. Nat Genet Vol. 2015;47:34.
Field Y, Boyle EA, Telis N, Gao Z, Gaulton KJ, Golan D, et al. Detection of human adaptation during the past 2000 years. Science. 2016;354:760–4.
Guo J, Wu Y, Zhu Z, Zheng Z, Trzaskowski M, Zeng J, et al. Global genetic differentiation of complex traits shaped by natural selection in humans. Nat Commun. 2018;9:1865.
Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–86.
Sohail M, Maier RM, Ganna A, Bloemendal A, Martin AR, Turchin MC, et al. Polygenic adaptation on height is overestimated due to uncorrected stratification in genome-wide association studies. Elife. 2019;8:1–17.
Berg JJ, Harpak A, Sinnott-Armstrong N, Joergensen AM, Mostafavi H, Field Y, et al. Reduced signal for polygenic adaptation of height in UK Biobank. Elife. 2019;8:1–47.
Chen M, Sidore C, Akiyama M, Ishigaki K, Kamatani Y, Schlessinger D, et al. Evidence of polygenic adaptation in Sardinia at height-associated loci ascertained from the Biobank Japan. Am J Hum Genet. 2020;107:60–71.
Tucci S, Vohr SH, McCoy RC, Vernot B, Robinson MR, Barbieri C, et al. Evolutionary history and adaptation of a human pygmy population of Flores Island, Indonesia. Science. 2018;361:511–6.
Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–91.
Mostafavi H, Harpak A, Agarwal I, Conley D, Pritchard JK, Przeworski M. Variable prediction accuracy of polygenic scores within an ancestry group. Elife. 2020;9:1–52.
1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Locke AE, Steinberg KM, Chiang CWK, Service SK, Havulinna AS, Stell L, et al. Exome sequencing of Finnish isolates enhances rare-variant association power. Nature. 2019;572:323–8.
Wang SR, Agarwala V, Flannick J, Chiang CWK, Altshuler D, Hirschhorn JN. Simulation of finnish population history, guided by empirical genetic data, to assess power of rare-variant tests in Finland. Am J Hum Genet. 2014;94:710–20.
Bergström A, McCarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, et al. Insights into human genetic variation and population history from 929 diverse genomes. Science. 2020;367:eaay5012.
Price AL, Weale ME, Patterson N, Myers SR, Need AC, Shianna KV, et al. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 2008;83:132–5.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Haller BC, Messer PW. SLiM 3: forward genetic simulations beyond the Wright-Fisher Model. Mol Biol Evol. 2019;36:632–7.
Kerminen S, Martin AR, Koskela J, Ruotsalainen SE, Havulinna AS, Surakka I, et al. Geographic variation and bias in the polygenic scores of complex diseases and traits in Finland. Am J Hum Genet. 2019;104:1169–81.
Edge MD, Coop G. Reconstructing the history of polygenic scores using coalescent trees. Genetics. 2019;211:235–62.
Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–24.
Mars N, Koskela JT, Ripatti P, Kiiskinen TTJ, Havulinna AS, Lindbohm JV, et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat Med. 2020;26:549–57.
Marnetto D, Pärna K, Läll K, Molinaro L, Montinaro F, Haller T, et al. Ancestry deconvolution and partial polygenic score can improve susceptibility predictions in recently admixed individuals. Nat Commun. 2020;11:1–9.
Dikilitas O, Schaid DJ, Kosel ML, Carroll RJ, Chute CG, Denny JA, et al. Predictive utility of polygenic risk scores for coronary heart disease in three major racial and ethnic groups. Am J Hum Genet. 2020;106:707–16.
Bitarello BD, Mathieson I. Polygenic scores for height in admixed populations. G3. 2020;10:4027–36.
Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100:635–49.
Barghi N, Hermisson J, Schlötterer C. Polygenic adaptation: a unifying framework to understand positive selection. Nat Rev Genet. 2020;21:769–81.
Kang SJ, Chiang CWK, Palmer CD, Tayo BO, Lettre G, Butler JL, et al. Genome-wide association of anthropometric traits in African- and African-derived populations. Hum Mol Genet. 2010;19:2725–38.
N’Diaye A, Chen GK, Palmer CD, Ge B, Tayo B, Mathias RA, et al. Identification, replication, and fine-mapping of loci associated with adult height in individuals of African ancestry. PLoS Genet. 2011;7:e1002298.
Akiyama M, Ishigaki K, Sakaue S, Momozawa Y, Horikoshi M, Hirata M, et al. Characterizing rare and low-frequency height-associated variants in the Japanese population. Nat Commun. 2019;10:4393.
Brick LA, Keller MC, Knopik VS, McGeary JE, Palmer RHC. Shared additive genetic variation for alcohol dependence among subjects of African and European ancestry. Addict Biol. 2019;24:132–44.
Zoledziewska M, Sidore C, Chiang CWK, Sanna S, Mulas A, Steri M, et al. Height-reducing variants and selection for short stature in Sardinia. Nat Genet. 2015;47:1352–6.
Hellenthal G, Busby GBJ, Band G, Wilson JF, Capelli C, Falush D, et al. A Genetic Atlas of Human Admixture History. Science. 2014;343:747–51.
Peter BM, Petkova D, Novembre J. Genetic landscapes reveal how human genetic diversity aligns with geography. Mol Biol Evol. 2020;37:943–51.
Botigué LR, Henn BM, Gravel S, Maples BK, Gignoux CR, Corona E, et al. Gene flow from North Africa contributes to differential human genetic diversity in southern europe. Proc Natl Acad Sci USA. 2013;110:11791–6.
Zaidi AA, Mathieson I. Demographic history mediates the effect of stratification on polygenic scores. eLife. 2020;9:e61548.
Mathieson I, McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat Genet. 2012;44:243–6.
Novembre J, Barton NH. Tread lightly interpreting polygenic tests of selection. Genetics. 2018;208:1351–5.
Acknowledgements
We gratefully thank Jing Guo and Jian Yang for providing the script for PS analysis. This work is supported by start-up funds provided by the Center for Genetic Epidemiology at the Keck School of Medicine of the University of Southern California (USC) (to CWKC). Computation for this work is supported by USC’s Center for Advanced Research Computing (https://carc.usc.edu).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Ethical approval is not required for this study, the data in this study are all publicly available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, M., Chiang, C.W.K. Allele frequency differentiation at height-associated SNPs among continental human populations. Eur J Hum Genet 29, 1542–1548 (2021). https://doi.org/10.1038/s41431-021-00938-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-021-00938-2