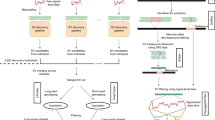
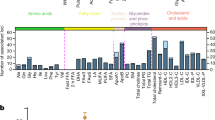
Abstract
We report ∼17.6 million genetic variants from whole-genome sequencing of 2,120 Sardinians; 22% are absent from previous sequencing-based compilations and are enriched for predicted functional consequences. Furthermore, ∼76,000 variants common in our sample (frequency >5%) are rare elsewhere (<0.5% in the 1000 Genomes Project). We assessed the impact of these variants on circulating lipid levels and five inflammatory biomarkers. We observe 14 signals, including 2 major new loci, for lipid levels and 19 signals, including 2 new loci, for inflammatory markers. The new associations would have been missed in analyses based on 1000 Genomes Project data, underlining the advantages of large-scale sequencing in this founder population.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Parkes, M. et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat. Genet. 39, 830–832 (2007).
Willer, C.J. et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 41, 25–34 (2009).
Chen, W. et al. Genetic variants near TIMP3 and high-density lipoprotein–associated loci influence susceptibility to age-related macular degeneration. Proc. Natl. Acad. Sci. USA 107, 7401–7406 (2010).
Do, R. et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat. Genet. 45, 1345–1352 (2013).
Do, R., Kathiresan, S. & Abecasis, G.R. Exome sequencing and complex disease: practical aspects of rare variant association studies. Hum. Mol. Genet. 21, R1–R9 (2012).
Zuk, O. et al. Searching for missing heritability: designing rare variant association studies. Proc. Natl. Acad. Sci. USA 111, E455–E464 (2014).
Kryukov, G.V., Shpunt, A., Stamatoyannopoulos, J.A. & Sunyaev, S.R. Power of deep, all-exon resequencing for discovery of human trait genes. Proc. Natl. Acad. Sci. USA 106, 3871–3876 (2009).
Peltonen, L., Palotie, A. & Lange, K. Use of population isolates for mapping complex traits. Nat. Rev. Genet. 1, 182–190 (2000).
Clarke, R. et al. Cholesterol fractions and apolipoproteins as risk factors for heart disease mortality in older men. Arch. Intern. Med. 167, 1373–1378 (2007).
Pai, J.K. et al. Inflammatory markers and the risk of coronary heart disease in men and women. N. Engl. J. Med. 351, 2599–2610 (2004).
Orrù, V. et al. Genetic variants regulating immune cell levels in health and disease. Cell 155, 242–256 (2013).
Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013).
Naitza, S. et al. A genome-wide association scan on the levels of markers of inflammation in Sardinians reveals associations that underpin its complex regulation. PLoS Genet. 8, e1002480 (2012).
Pilia, G. et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2, e132 (2006).
Sanna, S. et al. Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat. Genet. 42, 495–497 (2010).
Zoledziewska, M. et al. Variation within the CLEC16A gene shows consistent disease association with both multiple sclerosis and type 1 diabetes in Sardinia. Genes Immun. 10, 15–17 (2009).
Chen, W. et al. Genotype calling and haplotyping in parent-offspring trios. Genome Res. 23, 142–151 (2013).
Jun, G., Wing, M.K., Abecasis, G.R. & Kang, H.M. An efficient and scalable analysis framework for variant extraction and refinement from population scale DNA sequence data. Genome Res. 25, 918–925 (2015).
Li, Y., Sidore, C., Kang, H.M., Boehnke, M. & Abecasis, G.R. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 21, 940–951 (2011).
McLaren, W. et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26, 2069–2070 (2010).
Sherry, S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Francalacci, P. et al. Peopling of three Mediterranean islands (Corsica, Sardinia, and Sicily) inferred by Y-chromosome biallelic variability. Am. J. Phys. Anthropol. 121, 270–279 (2003).
Francalacci, P. et al. Low-pass DNA sequencing of 1200 Sardinians reconstructs European Y-chromosome phylogeny. Science 341, 565–569 (2013).
Zavattari, P. et al. Major factors influencing linkage disequilibrium by analysis of different chromosome regions in distinct populations: demography, chromosome recombination frequency and selection. Hum. Mol. Genet. 9, 2947–2957 (2000).
1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Novembre, J. et al. Genes mirror geography within Europe. Nature 456, 98–101 (2008).
Nelson, M.R. et al. The Population Reference Sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am. J. Hum. Genet. 83, 347–358 (2008).
Gravel, S. et al. Demographic history and rare allele sharing among human populations. Proc. Natl. Acad. Sci. USA 108, 11983–11988 (2011).
Nelson, M.R. et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 337, 100–104 (2012).
Mathieson, I. & McVean, G. Demography and the age of rare variants. PLoS Genet. 10, e1004528 (2014).
Chen, W.-M. & Abecasis, G.R. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 81, 913–926 (2007).
Li, Y., Willer, C., Sanna, S. & Abecasis, G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 10, 387–406 (2009).
Pistis, G. et al. Rare variant genotype imputation with thousands of study-specific whole-genome sequences: implications for cost-effective study designs. Eur. J. Hum. Genet. 23, 975–983 (2015).
Sanna, S. et al. Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet. 7, e1002198 (2011).
Cao, A. & Galanello, R. β-thalassemia. Genet. Med. 12, 61–76 (2010).
Maioli, M. et al. Plasma lipoprotein composition, apolipoprotein(a) concentration and isoforms in β-thalassemia. Atherosclerosis 131, 127–133 (1997).
Maioli, M. et al. Plasma lipids in β-thalassemia minor. Atherosclerosis 75, 245–248 (1989).
Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 46, 818–825 (2014).
Hou, S. et al. Genetic variant on PDGFRL associated with Behçet disease in Chinese Han populations. Hum. Mutat. 34, 74–78 (2013).
Xu, M. et al. An integrative approach to characterize disease-specific pathways and their coordination: a case study in cancer. BMC Genomics 9 (suppl. 1), S12 (2008).
Tournamille, C. et al. Arg89Cys substitution results in very low membrane expression of the Duffy antigen/receptor for chemokines in Fyx individuals. Blood 92, 2147–2156 (1998).
Shi, X.-F. et al. Structural analysis of human CCR2b and primate CCR2b by molecular modeling and molecular dynamics simulation. J. Mol. Model. 8, 217–222 (2002).
Schick, U.M. et al. Association of exome sequences with plasma C-reactive protein levels in >9000 participants. Hum. Mol. Genet. 24, 559–571 (2015).
Golledge, J. et al. Apolipoprotein E genotype is associated with serum C-reactive protein but not abdominal aortic aneurysm. Atherosclerosis 209, 487–491 (2010).
Kullo, I.J. et al. Complement receptor 1 gene variants are associated with erythrocyte sedimentation rate. Am. J. Hum. Genet. 89, 131–138 (2011).
Schadt, E.E. et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6, e107 (2008).
Yang, J., Lee, S.H., Goddard, M.E. & Visscher, P.M. Genome-wide complex trait analysis (GCTA): methods, data analyses, and interpretations. Methods Mol. Biol. 1019, 215–236 (2013).
Moltke, I. et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature 512, 190–193 (2014).
Danjou, F. et al. Genome-wide association analyses based on whole-genome sequencing in Sardinia provide insights into regulation of hemoglobin levels. Nat. Genet. doi: 10.1038/ng.3307 (14 September 2015).
Zoledziewska, M. et al. Height-reducing variants and selection for short stature in Sardinia. Nat. Genet. doi: 10.1038/ng.3403 (14 September 2015).
Pruim, R.J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Burdick, J.T., Chen, W.-M., Abecasis, G.R. & Cheung, V.G. In silico method for inferring genotypes in pedigrees. Nat. Genet. 38, 1002–1004 (2006).
Voight, B.F. et al. The Metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 8, e1002793 (2012).
Parkes, M., Cortes, A., van Heel, D.A. & Brown, M.A. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 14, 661–673 (2013).
Goldstein, J.I. et al. zCall: a rare variant caller for array-based genotyping: genetics and population analysis. Bioinformatics 28, 2543–2545 (2012).
McCarthy, M.I. et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9, 356–369 (2008).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Li, B. et al. QPLOT: a quality assessment tool for next generation sequencing data. BioMed. Res. Int. 2013, 865181 (2013).
Jun, G. et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am. J. Hum. Genet. 91, 839–848 (2012).
Li, Y., Willer, C.J., Ding, J., Scheet, P. & Abecasis, G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34, 816–834 (2010).
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. & Abecasis, G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959 (2012).
Price, A.L. et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 4, e236 (2008).
Lee, S., Zou, F. & Wright, F.A. Convergence and prediction of principal component scores in high-dimensional settings. Ann. Stat. 38, 3605–3629 (2010).
Kang, H.M. et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354 (2010).
Willer, C.J., Li, Y. & Abecasis, G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Li, B. & Leal, S.M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83, 311–321 (2008).
Price, A.L. et al. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 86, 832–838 (2010).
Zaitlen, N. et al. Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet. 9, e1003520 (2013).
Xu, C. et al. Estimating genome-wide significance for whole-genome sequencing studies. Genet. Epidemiol. 38, 281–290 (2014).
Abecasis, G.R., Cherny, S.S., Cookson, W.O. & Cardon, L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
Moayyeri, A., Hammond, C.J., Valdes, A.M. & Spector, T.D. Cohort profile: TwinsUK and Healthy Ageing Twin Study. Int. J. Epidemiol. 42, 76–85 (2013).
Esko, T. et al. Genetic characterization of northeastern Italian population isolates in the context of broader European genetic diversity. Eur. J. Hum. Genet. 21, 659–665 (2013).
Traglia, M. et al. Heritability and demographic analyses in the large isolated population of Val Borbera suggest advantages in mapping complex traits genes. PLoS ONE 4, e7554 (2009).
Winkelmann, B.R. et al. Rationale and design of the LURIC study—a resource for functional genomics, pharmacogenomics and long-term prognosis of cardiovascular disease. Pharmacogenomics 2, S1–S73 (2001).
Taylor, P.N. et al. Whole-genome sequence–based analysis of thyroid function. Nat. Commun. 6, 5681 (2015).
Acknowledgements
We thank all the volunteers who generously participated in this study and made this research possible. This research was supported by National Human Genome Research Institute grants HG005581, HG005552, HG006513, HG007022 and HG007089; by National Heart, Lung, and Blood Institute grant HL117626; by the Intramural Research Program of the US National Institutes of Health, National Institute on Aging, contracts N01-AG-1-2109 and HHSN271201100005C; by Sardinian Autonomous Region (L.R. 7/2009) grant cRP3-154; by the PB05 InterOmics MIUR Flagship Project; by grant FaReBio2011 'Farmaci e Reti Biotecnologiche di Qualità'; by a US National Institutes of Health National Research Service Award (NRSA) postdoctoral fellowship (F32GM106656) to C.W.K.C.; and by the UC MEXUS/CONOCYT fellowship to V.D.O.d.V. The replication cohorts acknowledge the use of data generated by the UK10K Consortium, supported by Wellcome Trust award WT091310. The UK10K research was specifically funded by a Wellcome Trust award, '10,000 UK Genome Sequences: Accessing the Role of Rare Genetic Variants in Health and Disease' (WT091310/C/10/Z). The research of N.S. is supported by the Wellcome Trust (grants WT098051 and WT091310), the European Union's Seventh Framework Programme (EPIGENESYS grant 257082 and BLUEPRINT grant HEALTH-F5-2011-282510) and the National Institute for Health Research (NIHR) British Research Council (BRC). The ING-FVG cohort was supported by grant Ministero della Salute—Ricerca Finalizzata PE-2011-02347500 (to P.G.); the ING-VB study thanks the inhabitants of Val Borbera for participating in the study, M. Traglia, C. Sala and C. Masciullo for data management, and the funding sources Fondazione Cariplo (Italy), the Ministry of Health, Ricerca Finalizzata (Italy) 2008, 2011-2012, and the Public Health Genomics Project 2010. The HELIC cohorts are thankful to the residents of the Pomak villages and the Mylopotamos villages for participating and to their funding sources, including the Wellcome Trust (098051) and the European Research Council (ERC-2011-StG 280559-SEPI).
Author information
Authors and Affiliations
Contributions
D.S., F.C. and G.R.A. conceived and supervised the study. C.S., S.N., S.S., D.S., F.C. and G.R.A. drafted the manuscript. E.P., M.Z., C.W.K.C. and J.N. revised the manuscript and wrote specific sections of it. F.B., A. Maschio, A.A., C.J. and R.L. supervised sequencing experiments. F.B., A. Maschio, B.T. and C.B. performed sequencing experiments. C.S., E.P., G.P., M.S., F.D. and S.S. carried out genetic association analyses. C.S., A.K., R.A., F.R., R.B., C.J., R.L. and H.M.K. were responsible for sequencing data processing. C.S., E.P. and G.P. analyzed DNA sequence data. M.Z., A. Mulas, F.B., S.U. and R.N. carried out SNP array genotyping. M.Z. designed the validation strategy, and M.Z., F.B. and A. Mulas verified genotypes by Sanger sequencing and TaqMan genotyping. C.S., J.B.-G., M.P., C.F. and S.S. were responsible for selection of samples for sequencing, J.N., C.W.K.C. and V.D.O.d.V. performed the allele-sharing, principal-component and FST analyses. J.H., P.G., G.M., N.J.T., E.Z., D.T., G.D. and N.S. provided replication results. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–10 and Supplementary Tables 1–17. (PDF 3248 kb)
Rights and permissions
About this article
Cite this article
Sidore, C., Busonero, F., Maschio, A. et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat Genet 47, 1272–1281 (2015). https://doi.org/10.1038/ng.3368
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3368
This article is cited by
-
Identifying the causal relationship between immune factors and osteonecrosis: a two-sample Mendelian randomization study
Scientific Reports (2024)
-
Causal relationships between peripheral immune cells and Alzheimer’s disease: a two-sample Mendelian randomization study
Neurological Sciences (2024)
-
Causal role of immune cells in schizophrenia: Mendelian randomization (MR) study
BMC Psychiatry (2023)
-
Genome-wide association study of varicose veins identifies a protective missense variant in GJD3 enriched in the Finnish population
Communications Biology (2023)
-
Polygenic risk score and biochemical/environmental variables predict a low-risk profile of age-related macular degeneration in Sardinia
Graefe's Archive for Clinical and Experimental Ophthalmology (2023)