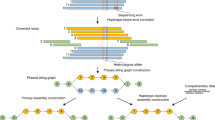
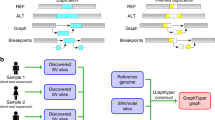
Abstract
Detecting genetic variants that are highly divergent from a reference sequence remains a major challenge in genome sequencing. We introduce de novo assembly algorithms using colored de Bruijn graphs for detecting and genotyping simple and complex genetic variants in an individual or population. We provide an efficient software implementation, Cortex, the first de novo assembler capable of assembling multiple eukaryotic genomes simultaneously. Four applications of Cortex are presented. First, we detect and validate both simple and complex structural variations in a high-coverage human genome. Second, we identify more than 3 Mb of sequence absent from the human reference genome, in pooled low-coverage population sequence data from the 1000 Genomes Project. Third, we show how population information from ten chimpanzees enables accurate variant calls without a reference sequence. Last, we estimate classical human leukocyte antigen (HLA) genotypes at HLA-B, the most variable gene in the human genome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Li, H., Ruan, J. & Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 (2008).
Li, R., Li, Y., Kristiansen, K. & Wang, J. SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714 (2008).
Lunter, G. & Goodson, M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21, 936–939 (2011).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Albers, C.A. et al. Dindel: accurate indel calls from short-read data. Genome Res. 21, 961–973 (2011).
Lee, S., Hormozdiari, F., Alkan, C. & Brudno, M. MoDIL: detecting small indels from clone-end sequencing with mixtures of distributions. Nat. Methods 6, 473–474 (2009).
Hajirasouliha, I. et al. Detection and characterization of novel sequence insertions using paired-end next-generation sequencing. Bioinformatics 26, 1277–1283 (2010).
Handsaker, R.E., Korn, J.M., Nemesh, J. & McCarroll, S.A. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat. Genet. 43, 269–276 (2011).
Korbel, J.O. et al. PEMer: a computational framework with simulation-based error models for inferring genomic structural variants from massive paired-end sequencing data. Genome Biol. 10, R23 (2009).
Korbel, J.O. et al. Paired-end mapping reveals extensive structural variation in the human genome. Science 318, 420–426 (2007).
Mills, R.E. et al. Mapping copy number variation by population-scale genome sequencing. Nature 470, 59–65 (2011).
Tuzun, E. et al. Fine-scale structural variation of the human genome. Nat. Genet. 37, 727–732 (2005).
Bentley, D.R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008).
Wang, J. et al. The diploid genome sequence of an Asian individual. Nature 456, 60–65 (2008).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Ge, F., Wang, L.S. & Kim, J. The cobweb of life revealed by genome-scale estimates of horizontal gene transfer. PLoS Biol. 3, e316 (2005).
Beiko, R.G., Harlow, T.J. & Ragan, M.A. Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. USA 102, 14332–14337 (2005).
Holcomb, C.L. et al. A multi-site study using high-resolution HLA genotyping by next generation sequencing. Tissue Antigens 77, 206–217 (2011).
Fonseca, V.G. et al. Second-generation environmental sequencing unmasks marine metazoan biodiversity. Nat. Commun. 1, 98 (2010).
Iafrate, A.J. et al. Detection of large-scale variation in the human genome. Nat. Genet. 36, 949–951 (2004).
Redon, R. et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006).
Sebat, J. et al. Large-scale copy number polymorphism in the human genome. Science 305, 525–528 (2004).
Sharp, A.J. et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 77, 78–88 (2005).
Kidd, J.M. et al. Mapping and sequencing of structural variation from eight human genomes. Nature 453, 56–64 (2008).
Myers, E.W. Toward simplifying and accurately formulating fragment assembly. J. Comput. Biol. 2, 275–290 (1995).
Myers, E.W. The fragment assembly string graph. Bioinformatics 21 (suppl. 2), ii79–ii85 (2005).
Simpson, J.T. & Durbin, R. Efficient construction of an assembly string graph using the FM-index. Bioinformatics 26, i367–i373 (2010).
Zerbino, D.R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Gnerre, S. et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA 108, 1513–1518 (2011).
Li, R. et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272 (2010).
Jones, T. et al. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 101, 7329–7334 (2004).
Vinson, J.P. et al. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi. Genome Res. 15, 1127–1135 (2005).
Kim, J.H., Waterman, M.S. & Li, L.M. Diploid genome reconstruction of Ciona intestinalis and comparative analysis with Ciona savignyi. Genome Res. 17, 1101–1110 (2007).
Donmez, N. & Brudno, M. Hapsembler: an assembler for highly polymorphic genomes. in Research in Computational Molecular Biology, Lecture Notes in Computer Science Vol. 6577 (eds. Bafna, V. & Sahinalp, S.), 38–52 (Springer, Berlin, Heidelberg, 2011).
Pevzner, P.A., Tang, H. & Waterman, M.S. An Eulerian path approach to DNA fragment assembly. Proc. Natl. Acad. Sci. USA 98, 9748–9753 (2001).
Idury, R.M. & Waterman, M.S. A new algorithm for DNA sequence assembly. J. Comput. Biol. 2, 291–306 (1995).
Simpson, J.T. et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 19, 1117–1123 (2009).
Zerbino, D.R., McEwen, G.K., Margulies, E.H. & Birney, E. Pebble and rock band: heuristic resolution of repeats and scaffolding in the velvet short-read de novo assembler. PLoS ONE 4, e8407 (2009).
Kidd, J.M. et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143, 837–847 (2010).
Myers, S. et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327, 876–879 (2010).
The International HapMap Consortium. et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
de Bakker, P.I. et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 38, 1166–1172 (2006).
Ratan, A., Yu, Z., Hayes, V.M., Schuster, S.C. & Miller, W. Calling SNPs without a reference sequence. BMC Bioinformatics 11, 130 (2010).
Peterlongo, P., Schnel, N., Pisanti, N., Sagot, M.-F. & Lacroix, V. Identifying SNPs without a reference genome by comparing raw reads. in String Processing and Information Retrieval—17th International Symposium (eds. Chavez, E. & Lonardi, S.) 147–158 (Los Cabos, Mexico, 2010).
Ding, L., Wendl, M.C., Koboldt, D.C. & Mardis, E.R. Analysis of next-generation genomic data in cancer: accomplishments and challenges. Hum. Mol. Genet. 19, R188–R196 (2010).
Harris, S.R. et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469–474 (2010).
Butler, J. et al. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 18, 810–820 (2008).
Chaisson, M.J., Brinza, D. & Pevzner, P.A. De novo fragment assembly with short mate-paired reads: does the read length matter? Genome Res. 19, 336–346 (2009).
Kelley, D.R., Schatz, M.C. & Salzberg, S.L. Quake: quality-aware detection and correction of sequencing errors. Genome Biol. 11, R116 (2010).
Allsopp, C.E. et al. Sequence analysis of HLA-Bw53, a common West African allele, suggests an origin by gene conversion of HLA-B35. Hum. Immunol. 30, 105–109 (1991).
Acknowledgements
We would like to thank the members of the 1000 Genomes Project Consortium for discussion, suggestions and sequencing data. We thank B. Ahiska, A. Auton, E. Birney, R. Durbin, G. Lunter, J. Woolf and D. Zerbino for discussion, two anonymous reviewers for their comments and members of the PanMap Project and the Genomics Core at the Wellcome Trust Centre for Human Genetics for access to sequence data. Z.I. is funded by a grant from the Wellcome Trust (WT086084/Z/08/Z to G.M.). The sequencing of NA12878 was performed by the Wellcome Trust Sequencing Core at Oxford, under a grant from the Wellcome Trust (090532/Z/09/Z).
Author information
Authors and Affiliations
Contributions
Z.I. and G.M. designed the study, developed the mathematical models and wrote the manuscript. M.C. and Z.I. developed the variant discovery algorithms, designed the multicolor graph data structures and implemented software. Z.I. performed simulations and analyses for cases 1, 3 and 4. I.T. and Z.I. performed analyses for case 2. P.F. contributed to early plans for Cortex.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–6 and Supplementary Tables 1–7 (PDF 1207 kb)
Rights and permissions
About this article
Cite this article
Iqbal, Z., Caccamo, M., Turner, I. et al. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat Genet 44, 226–232 (2012). https://doi.org/10.1038/ng.1028
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.1028
This article is cited by
-
Pan-genome de Bruijn graph using the bidirectional FM-index
BMC Bioinformatics (2023)
-
Detecting gene breakpoints in noisy genome sequences using position-annotated colored de-Bruijn graphs
BMC Bioinformatics (2023)
-
In vitro and in silico parameters for precise cgMLST typing of Listeria monocytogenes
BMC Genomics (2022)
-
Flexible protein database based on amino acid k-mers
Scientific Reports (2022)
-
Pangenome-based genome inference allows efficient and accurate genotyping across a wide spectrum of variant classes
Nature Genetics (2022)