Abstract
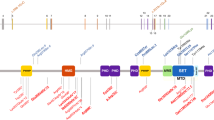
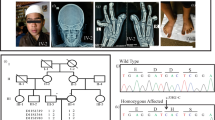
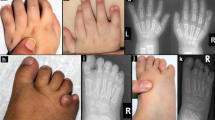
Myhre syndrome (MIM 139210) is a developmental disorder characterized by short stature, short hands and feet, facial dysmorphism, muscular hypertrophy, deafness and cognitive delay. Using exome sequencing of individuals with Myhre syndrome, we identified SMAD4 as a candidate gene that contributes to this syndrome on the basis of its pivotal role in the bone morphogenetic pathway (BMP) and transforming growth factor (TGF)-β signaling. We identified three distinct heterozygous missense SMAD4 mutations affecting the codon for Ile500 in 11 individuals with Myhre syndrome. All three mutations are located in the region of SMAD4 encoding the Mad homology 2 (MH2) domain near the site of monoubiquitination at Lys519, and we found a defect in SMAD4 ubiquitination in fibroblasts from affected individuals. We also observed decreased expression of downstream TGF-β target genes, supporting the idea of impaired TGF-β–mediated transcriptional control in individuals with Myhre syndrome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Myhre, S.A., Ruvalcaba, H.A. & Graham, C.B. A new growth deficiency syndrome. Clin. Genet. 20, 1–5 (1981).
Burglen, L. et al. Myhre syndrome: new reports, review, and differential diagnosis. J. Med. Genet. 40, 546–551 (2003).
Loeys, B.L. et al. Mutations in fibrillin-1cause congenital scleroderma: stiff skin syndrome. Sci. Transl. Med. 2, 23ra20 (2010).
Faivre, L. et al. Clinical homogeneity and genetic heterogeneity in Weill-Marchesani syndrome. Am. J. Med. Genet. 123, 204–207 (2003).
Le Goff, C. et al. Mutations in the TGF-β binding-protein-like domain 5 of FBN1 are responsible for acromicric and geleophysic dysplasias. Am. J. Hum. Genet. 89, 7–14 (2011).
Dagoneau, N. et al. ADAMTS10 mutations in autosomal recessive Weill-Marchesani syndrome. Am. J. Hum. Genet. 75, 801–806 (2004).
Le Goff, C. et al. ADAMTSL2 mutations in geleophysic dysplasia demonstrate a role for ADAMTS-like proteins in TGF-β bioavailability regulation. Nat. Genet. 40, 1119–1123 (2008).
Allali, S. et al. Molecular screening of ADAMTSL2 gene in 33 patients reveals the genetic heterogeneity of geleophysic dysplasia. J. Med. Genet. 48, 417–421 (2011).
Faivre, L. et al. In-frame fibrillin-1 gene deletion in autosomal dominant Weill-Marchesani syndrome. J. Med. Genet. 40, 34–36 (2003).
Ross, S. & Hill, C.S. How the Smads regulate transcription. Int. J. Biochem. Cell Biol. 40, 383–408 (2008).
Shi, Y. & Massague, J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700 (2003).
Sirard, C. et al. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later anterior development of the mouse embryo. Genes Dev. 12, 107–119 (1998).
Zhang, J. et al. Smad4 is required for the normal organization of the cartilage growth plate. Dev. Biol. 284, 311–322 (2005).
Yang, S.M. et al. Chondrocyte- specific Smad4 gene conditional knockout results in hearing loss and inner ear malformation in mice. Dev. Dyn. 238, 1897–1908 (2009).
Tan, X. et al. Smad4 is required for maintaining normal murine postnatal bone homeostasis. J. Cell Sci. 120, 2162–2170 (2007).
Howe, J.R. et al. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science 280, 1086–1088 (1998).
Schutte, M. et al. DPC4 gene in various tumor types. Cancer Res. 56, 2527–2530 (1996).
Hahn, S.A. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 271, 350–353 (1996).
Sunamura, M. et al. Gene therapy for pancreatic cancer based on genetic characterization of the disease. J. Hepatobiliary Pancreat. Surg. 9, 32–38 (2002).
Kurokawa, M. et al. The oncoprotein Evi-1 repress TGF-β signaling by inhibiting Smad3. Nature 394, 92–96 (1998).
Xu, J. & Attisano, L. Mutations in the tumor suppressors Smad2 and Smad4 inactivate transforming growth factor β signaling by targeting Smads to the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 97, 4820–4825 (2000).
Byun, M. et al. Whole-exome sequencing-based discovery of STIM1 deficiency in a child with fatal classic Kaposi sarcoma. J. Exp. Med. 207, 2307–2312 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Larrede, S. et al. Stimulation of cholesterol efflux by LXR agonists in cholesterol-loaded human macrophages is ABCA1-dependent but ABCG1-independent. Arterioscler. Thromb. Vasc. Biol. 29, 1930–1936 (2009).
Acknowledgements
We are grateful to the individuals with Myhre syndrome and their families for their participation in this study. We also thank the following physicians for the management of the affected individuals: D. Doummar, R. McGowan, P. Picco and M. Whiteford. This research was supported by the French National Research Agency (ANR; R09183KS to V.C.-D.).
Author information
Authors and Affiliations
Contributions
C.L.G. designed the experiments, analyzed the exome sequencing data, performed protein blot analysis and wrote the manuscript. C.M. performed Sanger sequencing analysis. A. Abhyankar and J.-L.C. performed exome capture. W.L.G. performed quantitative RT-PCR analysis. V.S. performed three-dimensional structure analysis. A. Afenjar, A.D., M.d.R., D.H., S.J., S.M., M.S., J.T. and A.V. provided clinical data. A.M. wrote the manuscript. V.C.-D. provided clinical data, analyzed the exome sequencing data, oversaw all aspects of the research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 and Supplementary Tables 1–4. (PDF 307 kb)
Rights and permissions
About this article
Cite this article
Le Goff, C., Mahaut, C., Abhyankar, A. et al. Mutations at a single codon in Mad homology 2 domain of SMAD4 cause Myhre syndrome. Nat Genet 44, 85–88 (2012). https://doi.org/10.1038/ng.1016
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.1016
This article is cited by
-
Natural history of Myhre syndrome
Orphanet Journal of Rare Diseases (2022)
-
Intervertebral disc degeneration is rescued by TGFβ/BMP signaling modulation in an ex vivo filamin B mouse model
Bone Research (2022)
-
Signaling network regulating osteogenesis in mesenchymal stem cells
Journal of Cell Communication and Signaling (2022)
-
Deficiency of TMEM53 causes a previously unknown sclerosing bone disorder by dysregulation of BMP-SMAD signaling
Nature Communications (2021)
-
SMAD4 mutations and cross-talk between TGF-β/IFNγ signaling accelerate rates of DNA damage and cellular senescence, resulting in a segmental progeroid syndrome—the Myhre syndrome
GeroScience (2021)