Abstract
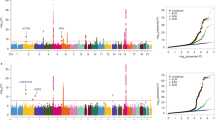
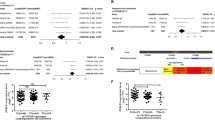
Lung cancer is the leading cause of cancer-related deaths worldwide. To identify genetic factors that modify the risk of lung cancer in individuals of Chinese ancestry, we performed a genome-wide association scan in 5,408 subjects (2,331 individuals with lung cancer (cases) and 3,077 controls) followed by a two-stage validation among 12,722 subjects (6,313 cases and 6,409 controls). The combined analyses identified six well-replicated SNPs with independent effects and significant lung cancer associations (P < 5.0 × 10−8) located in TP63 (rs4488809 at 3q28, P = 7.2 × 10−26), TERT-CLPTM1L (rs465498 and rs2736100 at 5p15.33, P = 1.2 × 10−20 and P = 1.0 × 10−27, respectively), MIPEP-TNFRSF19 (rs753955 at 13q12.12, P = 1.5 × 10−12) and MTMR3-HORMAD2-LIF (rs17728461 and rs36600 at 22q12.2, P = 1.1 × 10−11 and P = 6.2 × 10−13, respectively). Two of these loci (13q12.12 and 22q12.2) were newly identified in the Chinese population. These results suggest that genetic variants in 3q28, 5p15.33, 13q12.12 and 22q12.2 may contribute to the susceptibility of lung cancer in Han Chinese.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Zhang, H. & Cai, B. The impact of tobacco on lung health in China. Respirology 8, 17–21 (2003).
Hung, R.J. et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 452, 633–637 (2008).
Amos, C.I. et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 40, 616–622 (2008).
Thorgeirsson, T.E. et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 452, 638–642 (2008).
Wang, Y. et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet. 40, 1407–1409 (2008).
McKay, J.D. et al. Lung cancer susceptibility locus at 5p15.33. Nat. Genet. 40, 1404–1406 (2008).
Truong, T. et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J. Natl. Cancer Inst. 102, 959–971 (2010).
Broderick, P. et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 69, 6633–6641 (2009).
Wu, C. et al. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res. 69, 5065–5072 (2009).
Miki, D. et al. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat. Genet. 42, 893–896 (2010).
Landi, M.T. et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 85, 679–691 (2009).
Yamamoto, K., Okamoto, A., Isonishi, S., Ochiai, K. & Ohtake, Y. A novel gene, CRR9, which was up-regulated in CDDP-resistant ovarian tumor cell line, was associated with apoptosis. Biochem. Biophys. Res. Commun. 280, 1148–1154 (2001).
Chew, A., Sirugo, G., Alsobrook, J.P. II & Isaya, G. Functional and genomic analysis of the human mitochondrial intermediate peptidase, a putative protein partner of frataxin. Genomics 65, 104–112 (2000).
Eby, M.T., Jasmin, A., Kumar, A., Sharma, K. & Chaudhary, P.M. TAJ, a novel member of the tumor necrosis factor receptor family, activates the c-Jun N-terminal kinase pathway and mediates caspase-independent cell death. J. Biol. Chem. 275, 15336–15342 (2000).
Bei, J.X. et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat. Genet. 42, 599–603 (2010).
Niwa, H., Ogawa, K., Shimosato, D. & Adachi, K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460, 118–122 (2009).
Chen, Y. et al. Gprc5a deletion enhances the transformed phenotype in normal and malignant lung epithelial cells by eliciting persistent Stat3 signaling induced by autocrine leukemia inhibitory factor. Cancer Res. 70, 8917–8926 (2010).
Yu, H., Pardoll, D. & Jove, R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9, 798–809 (2009).
Zheng, L. et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat. Med. 13, 812–819 (2007).
Li, M. et al. XRCC1 polymorphisms, cooking oil fume and lung cancer in Chinese women nonsmokers. Lung Cancer 62, 145–151 (2008).
Lu, J. et al. Functional characterization of a promoter polymorphism in APE1/Ref-1 that contributes to reduced lung cancer susceptibility. FASEB J. 23, 3459–3469 (2009).
Guo, H. et al. Functional promoter -1271G>C variant of HSPB1 predicts lung cancer risk and survival. J. Clin. Oncol. 28, 1928–1935 (2010).
Liu, B. et al. A functional variant (-1304T>G) in the MKK4 promoter contributes to a decreased risk of lung cancer by increasing the promoter activity. Carcinogenesis 31, 1405–1411 (2010).
Purdue, M.P. et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat. Genet. 43, 60–65 (2011).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Barrett, J.C. et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Stephens, M. et al. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (2001).
Acknowledgements
This work is funded by the China National High-Tech Research and Development Program Grant (2009AA022705, 2009AA022701) and partly funded by the National Key Basic Research Program Grant (2011CB503805), the National Natural Science Foundation of China (30730080, 30972541, 30901233 and 30872178) and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions. The authors wish to thank all the study participants, research staff and students who participated in this work. We thank C. Amos, J. Xu and Q. Wei for their manuscript editing.
Author information
Authors and Affiliations
Contributions
H.S. directed the study, obtained financial support and was responsible for study design, interpretation of results and manuscript writing. Z.H. performed overall project management, statistical analyses with J.D. and drafted the initial manuscript. D. Lin, T.W., Y.S., D. Lu., J.L., B.Z., X.W. and L.J. directed each participating study and jointly organized this study. L.H., Y.J., M.C., J.C. and F.L. were responsible for sample processing and managing the genotyping data. Y.C., Y.S., L.X., G.J., Z.Z. and H.M. were responsible for subject recruitment and sample preparation of Nanjing samples. X.L., C.W., W.T. and D.Y. were responsible for subject recruitment and sample preparation of Beijing samples. L.L., P.X., H.G. and Q.D. were responsible for subject recruitment and sample preparation of Wuhan samples. B.H., C.B., Z.L., X.Z., G.Z., J.W. and H.C. were responsible for subject recruitment and sample preparation of Shanghai samples. Y.Z., H.Z., Y.Y., Z.Y., W.W. and P.G. were responsible for subject recruitment and sample preparation of Shenyang samples. L.Y. was responsible for subject recruitment and sample preparation of Guangzhou samples. F.C. oversaw the statistical analyses process. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Tables 1–8. (PDF 1068 kb)
Rights and permissions
About this article
Cite this article
Hu, Z., Wu, C., Shi, Y. et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet 43, 792–796 (2011). https://doi.org/10.1038/ng.875
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.875
This article is cited by
-
Contribution of an Asian-prevalent HLA haplotype to the risk of HBV-related hepatocellular carcinoma
Scientific Reports (2023)
-
Ethnicity-specific association between TERT rs2736100 (A > C) polymorphism and lung cancer risk: a comprehensive meta-analysis
Scientific Reports (2023)
-
Integrative analysis of GWAS and transcriptomics data reveal key genes for non-small lung cancer
Medical Oncology (2023)
-
Genome-wide association study of lung adenocarcinoma in East Asia and comparison with a European population
Nature Communications (2023)
-
TERT Gene rs2736100 and rs2736098 Polymorphisms are Associated with Increased Cancer Risk: A Meta-Analysis
Biochemical Genetics (2022)