Abstract
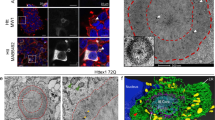
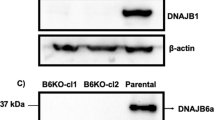
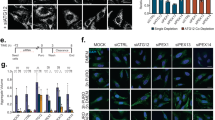
Continuous turnover of intracellular components by autophagy is necessary to preserve cellular homeostasis in all tissues. Alterations in macroautophagy, the main process responsible for bulk autophagic degradation, have been proposed to contribute to pathogenesis in Huntington's disease (HD), a genetic neurodegenerative disorder caused by an expanded polyglutamine tract in the huntingtin protein. However, the precise mechanism behind macroautophagy malfunction in HD is poorly understood. In this work, using cellular and mouse models of HD and cells from humans with HD, we have identified a primary defect in the ability of autophagic vacuoles to recognize cytosolic cargo in HD cells. Autophagic vacuoles form at normal or even enhanced rates in HD cells and are adequately eliminated by lysosomes, but they fail to efficiently trap cytosolic cargo in their lumen. We propose that inefficient engulfment of cytosolic components by autophagosomes is responsible for their slower turnover, functional decay and accumulation inside HD cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Morimoto, R.I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22, 1427–1438 (2008).
Rubinsztein, D.C. Lessons from animal models of Huntington's disease. Trends Genet. 18, 202–209 (2002).
Sarkar, S. & Rubinsztein, D.C. Huntington's disease: degradation of mutant huntingtin by autophagy. FEBS J. 275, 4263–4270 (2008).
Shibata, M. et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J. Biol. Chem. 281, 14474–14485 (2006).
Jeong, H. et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell 137, 60–72 (2009).
Iwata, A. et al. Intra-nuclear degradation of polyglutamine aggregates by the ubiquitin proteasome system. J. Biol. Chem. 284, 9796–9803 (2009).
Mizushima, N., Levine, B., Cuervo, A. & Klionsky, D. Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008).
Ravikumar, B., Duden, R. & Rubinsztein, D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 11, 1107–1117 (2002).
Ravikumar, B. et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36, 585–595 (2004).
Sarkar, S., Davies, J.E., Huang, Z., Tunnacliffe, A. & Rubinsztein, D.C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 282, 5641–5652 (2007).
Kegel, K.B. et al. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J. Neurosci. 20, 7268–7278 (2000).
Sapp, E. et al. Huntingtin localization in brains of normal and Huntington's disease patients. Ann. Neurol. 42, 604–612 (1997).
Davies, S.W. et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90, 537–548 (1997).
Nagata, E., Sawa, A., Ross, C.A. & Snyder, S.H. Autophagosome-like vacuole formation in Huntington's disease lymphoblasts. Neuroreport 15, 1325–1328 (2004).
Atwal, R.S. et al. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet. 16, 2600–2615 (2007).
Kim, J., Huang, W.P., Stromhaug, P.E. & Klionsky, D.J. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 277, 763–773 (2002).
Ravikumar, B., Imarisio, S., Sarkar, S., O'Kane, C.J. & Rubinsztein, D.C. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J. Cell Sci. 121, 1649–1660 (2008).
Wheeler, V.C. et al. Length-dependent gametic CAG repeat instability in the Huntington's disease knock-in mouse. Hum. Mol. Genet. 8, 115–122 (1999).
Klionsky, D.J., Cuervo, A.M. & Seglen, P.O Methods for monitoring autophagy from yeast to human. Autophagy 3, 181–206 (2007).
Trettel, F. et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet. 9, 2799–2809 (2000).
Kabeya, Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000).
Marzella, L., Ahlberg, J. & Glaumann, H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J. Cell Biol. 93, 144–154 (1982).
Kim, P.K., Hailey, D.W., Mullen, R.T. & Lippincott-Schwartz, J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. USA 105, 20567–20574 (2008).
Filimonenko, M. et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179, 485–500 (2007).
Singh, R. et al. Autophagy regulates lipid metabolism. Nature 458, 1131–1135 (2009).
Browne, S.E., Ferrante, R.J. & Beal, M.F. Oxidative stress in Huntington's disease. Brain Pathol. 9, 147–163 (1999).
Larsen, K.E., Fon, E.A., Hastings, T.G., Edwards, R.H. & Sulzer, D. Methamphetamine-induced degeneration of dopaminergic neurons involves autophagy and upregulation of dopamine synthesis. J. Neurosci. 22, 8951–8960 (2002).
Rubinsztein, D.C. et al. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy 1, 11–22 (2005).
Chu, C.T. et al. Autophagy in neurite injury and neurodegeneration: in vitro and in vivo models. Methods Enzymol. 453, 217–249 (2009).
Wang, Y. et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum. Mol. Genet. 18, 4153–4170 (2009).
Cuervo, A.M., Stefanis, L., Fredenburg, R., Lansbury, P.T. & Sulzer, D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305, 1292–1295 (2004).
Stefanis, L., Larsen, K., Rideout, H., Sulzer, D. & Greene, L. Expression of A53T mutant but not wild-type alpha-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J. Neurosci. 21, 9549–9560 (2001).
Yu, W.H. et al. Macroautophagy–a novel Beta-amyloid peptide-generating pathway activated in Alzheimer's disease. J. Cell Biol. 171, 87–98 (2005).
Sulzer, D. et al. Neuromelanin biosynthesis is driven by excess cytosolic catecholamines not accumulated by synaptic vesicles. Proc. Natl. Acad. Sci. USA 97, 11869–11874 (2000).
Bae, B.I. et al. Mutant huntingtin: nuclear translocation and cytotoxicity mediated by GAPDH. Proc. Natl. Acad. Sci. USA 103, 3405–3409 (2006).
Browne, S.E. Mitochondria and Huntington's disease pathogenesis: insight from genetic and chemical models. Ann. NY Acad. Sci. 1147, 358–382 (2008).
Wang, H., Lim, P.J., Karbowski, M. & Monteiro, M.J. Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum. Mol. Genet. 18, 737–752 (2009).
Twig, G. et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446 (2008).
Martin-Aparicio, E. et al. Proteasomal-dependent aggregate reversal and absence of cell death in a conditional mouse model of Huntington's disease. J. Neurosci. 21, 8772–8781 (2001).
Garfield, A.S. Derivation of primary mouse embryonic fibroblast (PMEF) cultures. Methods Mol. Biol. 633, 19–27 (2010).
Petersen, A. et al. Expanded CAG repeats in exon 1 of the Huntington's disease gene stimulate dopamine-mediated striatal neuron autophagy and degeneration. Hum. Mol. Genet. 10, 1243–1254 (2001).
Massey, A.C., Kaushik, S., Sovak, G., Kiffin, R. & Cuervo, A.M. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA 103, 5805–5810 (2006).
Klionsky, D.J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175 (2008).
Ohsumi, Y., Ishikawa, T. & Kato, K. A rapid and simplified method for the preparation of lysosomal membranes from rat liver. J. Biochem. 93, 547–556 (1983).
Storrie, B. & Madden, E. Isolation of subcellular organelles. Methods Enzymol. 182, 203–225 (1990).
Acknowledgements
We thank R. Singh and B. Patel for technical assistance. Mouse fibroblasts expressing 146Q-htt exon 1 and human lymphoblasts from unaffected controls and individuals with HD were gifts from N. Wexler (Hereditary Disease Foundation), M. Andresen (Massachusetts Institute of Technology) and J. Gusella (Massachusetts General Hospital). The antibodies to mannose-6-phosphate receptor and Rab5 were a gift from A. Wolkoff (Albert Einstein College of Medicine). This work was supported by a Huntington Disease Society of America grant (D.S., A.M.C.); US National Institutes of Health National Institute on Aging grants AG021904, AG031782, DK041918 (A.M.C.) and National Institute of Neurological Disorders and Stroke Udall Center of Excellence grant (D.S.); and the Glenn Foundation (A.M.C.), and Picower and Simons foundation (D.S.). E.W. is supported by the Hereditary Disease Foundation. E.A. is supported by a Fulbright fellowship. S.K. is supported by US National Institutes of Health National Institute on Aging grant T32AG023475.
Author information
Authors and Affiliations
Contributions
M.M.-V., Z.T. and E.W. performed the experiments that constitute the main body of this work; R.d.V., H.K., S.K., E.A. and G.T. completed the rest of the experiments; S.H. analyzed the electron micrographs of lymphoblasts; A.M.C. and D.S. designed the study and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–21 and Supplementary Text (PDF 3527 kb)
Rights and permissions
About this article
Cite this article
Martinez-Vicente, M., Talloczy, Z., Wong, E. et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci 13, 567–576 (2010). https://doi.org/10.1038/nn.2528
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.2528
This article is cited by
-
Upregulated pexophagy limits the capacity of selective autophagy
Nature Communications (2024)
-
Spotlights on ubiquitin-specific protease 12 (USP12) in diseases: from multifaceted roles to pathophysiological mechanisms
Journal of Translational Medicine (2023)
-
The regulatory role of lipophagy in central nervous system diseases
Cell Death Discovery (2023)
-
Mitophagy regulation in aging and neurodegenerative disease
Biophysical Reviews (2023)
-
Targeting Sigma Receptors for the Treatment of Neurodegenerative and Neurodevelopmental Disorders
CNS Drugs (2023)