Abstract
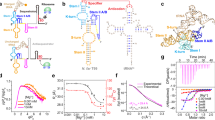
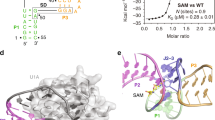
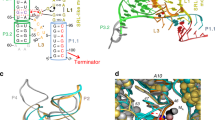
S-adenosylmethionine (SAM) riboswitches are widespread in bacteria, and up to five different SAM riboswitch families have been reported, highlighting the relevance of SAM regulation. On the basis of crystallographic and biochemical data, it has been postulated, but never demonstrated, that ligand recognition by SAM riboswitches involves key conformational changes in the RNA architecture. We show here that the aptamer follows a two-step hierarchical folding selectively induced by metal ions and ligand binding, each of them leading to the formation of one of the two helical stacks observed in the crystal structure. Moreover, we find that the anti-antiterminator P1 stem is rotated along its helical axis upon ligand binding, a mechanistic feature that could be common to other riboswitches. We also show that the nonconserved P4 helical domain is used as an auxiliary element to enhance the ligand-binding affinity. This work provides the first comprehensive characterization, to our knowledge, of a ligand-controlled riboswitch folding pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Serganov, A. & Patel, D.J. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 8, 776–790 (2007).
Roth, A. & Breaker, R.R. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 78, 305–334 (2009).
Loh, E. et al. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139, 770–779 (2009).
Schwalbe, H., Buck, J., Furtig, B., Noeske, J. & Wohnert, J. Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew. Chem. Int. Edn Engl. 46, 1212–1219 (2007).
Edwards, T.E., Klein, D.J. & Ferre-D'Amare, A.R. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr. Opin. Struct. Biol. 17, 273–279 (2007).
Dambach, M.D. & Winkler, W.C. Expanding roles for metabolite-sensing regulatory RNAs. Curr. Opin. Microbiol. 12, 161–169 (2009).
Cromie, M.J., Shi, Y., Latifi, T. & Groisman, E.A. An RNA sensor for intracellular Mg(2+). Cell 125, 71–84 (2006).
Nechooshtan, G., Elgrably-Weiss, M., Sheaffer, A., Westhof, E. & Altuvia, S. A pH-responsive riboregulator. Genes Dev. 23, 2650–2662 (2009).
Chowdhury, S., Maris, C., Allain, F.H. & Narberhaus, F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 25, 2487–2497 (2006).
Grundy, F.J. & Henkin, T.M. Regulation of gene expression by effectors that bind to RNA. Curr. Opin. Microbiol. 7, 126–131 (2004).
Kim, J.N. et al. Design and antimicrobial action of purine analogues that bind Guanine riboswitches. ACS Chem. Biol. 4, 915–927 (2009).
Mulhbacher, J. et al. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog. 6, e1000865 (2010).
Mulhbacher, J., St-Pierre, P. & Lafontaine, D.A. Therapeutic applications of ribozymes and riboswitches. Curr. Opin. Pharmacol. 10, 551–556 (2010).
Wang, J.X., Lee, E.R., Morales, D.R., Lim, J. & Breaker, R.R. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol. Cell 29, 691–702 (2008).
McDaniel, B.A., Grundy, F.J., Artsimovitch, I. & Henkin, T.M. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. USA 100, 3083–3088 (2003).
Winkler, W.C., Nahvi, A., Sudarsan, N., Barrick, J.E. & Breaker, R.R. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 10, 701–707 (2003).
Montange, R.K. & Batey, R.T. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441, 1172–1175 (2006).
Heppell, B. & Lafontaine, D.A. Folding of the SAM aptamer is determined by the formation of a K-turndependent pseudoknot. Biochemistry 47, 1490–1499 (2008).
McDaniel, B.A., Grundy, F.J. & Henkin, T.M. A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine-directed transcription termination. Mol. Microbiol. 57, 1008–1021 (2005).
Montange, R.K. & Batey, R.T. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys 37, 117–133 (2008).
Baird, N.J. & Ferre-D'Amare, A.R. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA 16, 598–609 (2010).
Epshtein, V., Mironov, A.S. & Nudler, E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. USA 100, 5052–5056 (2003).
Greenleaf, W.J., Frieda, K.L., Foster, D.A., Woodside, M.T. & Block, S.M. Direct observation of hierarchical folding in single riboswitch aptamers. Science 319, 630–633 (2008).
Lilley, D.M. Analysis of global conformation of branched RNA species using electrophoresis and fluorescence. Methods Enzymol. 317, 368–393 (2000).
Lumpkin, O.J. Mobility of DNA in gel electrophoresis. Biopolymers 21, 2315–2316 (1982).
Roy, R., Hohng, S. & Ha, T. A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 (2008).
Weinberg, Z. et al. The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches. RNA 14, 822–828 (2008).
Merino, E.J., Wilkinson, K.A., Coughlan, J.L. & Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127, 4223–4231 (2005).
Huang, W., Kim, J., Jha, S. & Aboul-ela, F. A mechanism for S-adenosyl methionine assisted formation of a riboswitch conformation: a small molecule with a strong arm. Nucleic Acids Res. 37, 6528–6539 (2009).
Garst, A.D., Heroux, A., Rambo, R.P. & Batey, R.T. Crystal structure of the lysine riboswitch regulatory mRNA element. J. Biol. Chem. 283, 22347–22351 (2008).
Lemay, J.F., Penedo, J.C., Tremblay, R., Lilley, D.M. & Lafontaine, D.A. Folding of the adenine riboswitch. Chem. Biol. 13, 857–868 (2006).
Noeske, J. et al. Interplay of “induced fit” and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 35, 572–583 (2007).
Stoddard, C.D., Gilbert, S.D. & Batey, R.T. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA 14, 675–684 (2008).
Lang, K., Rieder, R. & Micura, R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 35, 5370–5378 (2007).
Brenner, M.D., Scanlan, M.S., Nahas, M.K., Ha, T. & Silverman, S.K. Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine. Biochemistry 49, 1596–1605 (2010).
Lipfert, J. et al. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J. Mol. Biol. 365, 1393–1406 (2007).
Stoddard, C.D. et al. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure 18, 787–797 (2010).
Hennelly, S.P. & Sanbonmatsu, K.Y. Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Res. 39, 2416–2431 (2011).
Gilbert, S.D., Love, C.E., Edwards, A.L. & Batey, R.T. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry 46, 13297–13309 (2007).
Delfosse, V. et al. Riboswitch structure: an internal residue mimicking the purine ligand. Nucleic Acids Res. 38, 2057–2068 (2010).
Blouin, S., Chinnappan, R. & Lafontaine, D.A. Folding of the lysine riboswitch: importance of peripheral elements for transcriptional regulation. Nucleic Acids Res., doi:10.1093/nar/gkq1247 (published online 17 December 2010).
Mahen, E.M., Watson, P.Y., Cottrell, J.W. & Fedor, M.J. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol. 8, e1000307 (2010).
Whitford, P.C. et al. Nonlocal helix formation is key to understanding S-adenosylmethionine-1 riboswitch function. Biophys. J. 96, L7–L9 (2009).
Tomsic, J., McDaniel, B.A., Grundy, F.J. & Henkin, T.M. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in Bacillus subtilis exhibit differential sensitivity to SAM in vivo and in vitro. J. Bacteriol. 190, 823–833 (2008).
Wickiser, J.K., Cheah, M.T., Breaker, R.R. & Crothers, D.M. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry 44, 13404–13414 (2005).
Wickiser, J.K., Winkler, W.C., Breaker, R.R. & Crothers, D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell 18, 49–60 (2005).
Pan, T. & Sosnick, T. RNA folding during transcription. Annu. Rev. Biophys. Biomol. Struct. 35, 161–175 (2006).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brunger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54, 905–921 (1998).
Acknowledgements
We thank A. Lavigueur for critical reading of the manuscript. We also thank D. Norman and P. St.-Pierre for discussion. This work was supported by the Canadian Institutes of Health Research (CIHR). S.B. is supported by a studentship from the National Sciences and Engineering Research Council of Canada. D.A.L. is a CIHR New Investigator scholar as well as a Chercheur-boursier Junior 2 from the Fonds de la recherche en Santé du Québec. J.C.P. thanks the UK Biological and Biotechnological Science Research Council. We are also grateful to T. Ha and S. McKinney (University of Illinois at Urbana-Champaign) for providing the source code software for hidden Markov analysis.
Author information
Authors and Affiliations
Contributions
B.H. performed experiments, analyzed data and wrote the paper. S.B. conducted chemical probing. A-M.D. and J.M. carried out comparative gel electrophoresis and in vitro transcription controls, respectively. E.E. performed molecular modeling. J.C.P. and D.A.L. analyzed data and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Figures 1–19 (PDF 18526 kb)
Rights and permissions
About this article
Cite this article
Heppell, B., Blouin, S., Dussault, AM. et al. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol 7, 384–392 (2011). https://doi.org/10.1038/nchembio.563
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.563
This article is cited by
-
Exploring the energy landscape of a SAM-I riboswitch
Journal of Biological Physics (2021)
-
Development of a genetically encodable FRET system using fluorescent RNA aptamers
Nature Communications (2018)
-
Light-activated chemical probing of nucleobase solvent accessibility inside cells
Nature Chemical Biology (2018)
-
An excited state underlies gene regulation of a transcriptional riboswitch
Nature Chemical Biology (2017)
-
Single-molecule FRET reveals the energy landscape of the full-length SAM-I riboswitch
Nature Chemical Biology (2017)