Abstract
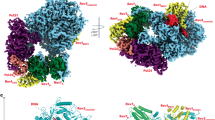
The Varkud satellite (VS) ribozyme mediates rolling-circle replication of a plasmid found in the Neurospora mitochondrion. We report crystal structures of this ribozyme from Neurospora intermedia at 3.1 Å resolution, which revealed an intertwined dimer formed by an exchange of substrate helices. In each protomer, an arrangement of three-way helical junctions organizes seven helices into a global fold that creates a docking site for the substrate helix of the other protomer, resulting in the formation of two active sites in trans. This mode of RNA−RNA association resembles the process of domain swapping in proteins and has implications for RNA regulation and evolution. Within each active site, adenine and guanine nucleobases abut the scissile phosphate, poised to serve direct roles in catalysis. Similarities to the active sites of the hairpin and hammerhead ribozymes highlight the functional importance of active-site features, underscore the ability of RNA to access functional architectures from distant regions of sequence space, and suggest convergent evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lilley, D.M.J. & Eckstein, F. Ribozymes and RNA Catalysis (eds. Lilley, D.M.J. & Eckstein, F.) 1–7 (RSC Publishing, Cambridge, 2008).
Thompson, J.E. & Raines, R.T. Value of general acid-base catalysis to ribonuclease A. J. Am. Chem. Soc. 116, 5467–5468 (1994).
Cochrane, J.C. & Strobel, S.A. Catalytic strategies of self-cleaving ribozymes. Acc. Chem. Res. 41, 1027–1035 (2008).
Kennell, J.C. et al. The VS catalytic RNA replicates by reverse transcription as a satellite of a retroplasmid. Genes Dev. 9, 294–303 (1995).
Saville, B.J. & Collins, R.A. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell 61, 685–696 (1990).
Beattie, T.L., Olive, J.E. & Collins, R.A. A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc. Natl. Acad. Sci. USA 92, 4686–4690 (1995).
Jones, F.D., Ryder, S.P. & Strobel, S.A. An efficient ligation reaction promoted by a Varkud Satellite ribozyme with extended 5′- and 3′- termini. Nucleic Acids Res. 29, 5115–5120 (2001).
Guo, H.C. & Collins, R.A. Efficient trans-cleavage of a stem-loop RNA substrate by a ribozyme derived from Neurospora VS RNA. EMBO J. 14, 368–376 (1995).
Ouellet, J., Byrne, M. & Lilley, D.M. Formation of an active site in trans by interaction of two complete Varkud Satellite ribozymes. RNA 15, 1822–1826 (2009).
Kath-Schorr, S. et al. General acid-base catalysis mediated by nucleobases in the hairpin ribozyme. J. Am. Chem. Soc. 134, 16717–16724 (2012).
Wilson, T.J., McLeod, A.C. & Lilley, D.M. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 26, 2489–2500 (2007).
Sood, V.D. & Collins, R.A. Identification of the catalytic subdomain of the VS ribozyme and evidence for remarkable sequence tolerance in the active site loop. J. Mol. Biol. 320, 443–454 (2002).
Lafontaine, D.A., Wilson, T.J., Zhao, Z.Y. & Lilley, D.M. Functional group requirements in the probable active site of the VS ribozyme. J. Mol. Biol. 323, 23–34 (2002).
Wilson, T.J. & Lilley, D.M.J. Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid-base catalysis? A critical assessment of available experimental data. RNA 17, 213–221 (2011).
Rupert, P.B. & Ferré-D'Amaré, A.R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410, 780–786 (2001).
Andersen, A.A. & Collins, R.A. Intramolecular secondary structure rearrangement by the kissing interaction of the Neurospora VS ribozyme. Proc. Natl. Acad. Sci. USA 98, 7730–7735 (2001).
Andersen, A.A. & Collins, R.A. Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol. Cell 5, 469–478 (2000).
Zamel, R. & Collins, R.A. Rearrangement of substrate secondary structure facilitates binding to the Neurospora VS ribozyme. J. Mol. Biol. 324, 903–915 (2002).
Koldobskaya, Y. et al. A portable RNA sequence, whose recognition by a synthetic antibody facilitates structural determination. Nat. Struct. Mol. Biol. 18, 100–106 (2011).
Rousseau, F., Schymkowitz, J. & Itzhaki, L.S. Implications of 3D domain swapping for protein folding, misfolding and function. Adv. Exp. Med. Biol. 747, 137–152 (2012).
Lafontaine, D.A., Norman, D.G. & Lilley, D.M. The global structure of the VS ribozyme. EMBO J. 21, 2461–2471 (2002).
Lipfert, J., Ouellet, J., Norman, D.G., Doniach, S. & Lilley, D.M. The complete VS ribozyme in solution studied by small-angle X-ray scattering. Structure 16, 1357–1367 (2008).
Rastogi, T., Beattie, T.L., Olive, J.E. & Collins, R.A. A long range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J. 15, 2820–2825 (1996).
Bouchard, P. et al. Role of SLV in SLI substrate recognition by the Neurospora VS ribozyme. RNA 14, 736–748 (2008).
Bouchard, P. & Legault, P. A remarkably stable kissing-loop interaction defines substrate recognition by the Neurospora Varkud Satellite ribozyme. RNA 20, 1451–1464 (2014).
Suslov, N.B. Crystal Structure of the VS Ribozyme: Implications for RNA Catalysis, Biology and Evolution. Thesis, University of Chicago (2012).
Poon, A.H.L., Olive, J.E., McLaren, M. & Collins, R.A. Identification of separate structural features that affect rate and cation concentration dependence of self-cleavage by the Neurospora VS ribozyme. Biochemistry 45, 13394–13400 (2006).
Bonneau, E. & Legault, P. Nuclear magnetic resonance structure of the III–IV-V three-way junction from the Varkud Satellite ribozyme and identification of magnesium-binding sites using paramagnetic relaxation enhancement. Biochemistry 53, 6264–6275 (2014).
Lafontaine, D.A., Norman, D.G. & Lilley, D.M. Structure, folding and activity of the VS ribozyme: importance of the 2–3-6 helical junction. EMBO J. 20, 1415–1424 (2001).
Hiley, S.L., Sood, V.D., Fan, J. & Collins, R.A. 4-thio-U cross-linking identifies the active site of the VS ribozyme. EMBO J. 21, 4691–4698 (2002).
Liu, Y., Wilson, T.J., McPhee, S. & Lilley, D.M.J. Crystal structure and mechanistic investigation of the Twister ribozyme. Nat. Chem. Biol. 10, 739–744 (2014).
Eiler, D., Wang, J. & Steitz, T.A. Structural basis for the fast self-cleavage reaction catalyzed by the twister ribozyme. Proc. Natl. Acad. Sci. USA 111, 13028–13033 (2014).
Ren, A. et al. In-line alignment and Mg2+ coordination at the cleavage site of the env22 twister ribozyme. Nat. Commun. 5, 5534 (2014).
Wilson, T.J. et al. Nucleobase-mediated general acid-base catalysis in the Varkud satellite ribozyme. Proc. Natl. Acad. Sci. USA 107, 11751–11756 (2010).
Pinard, R. et al. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 20, 6334–6442 (2001).
Heldenbrand, H. et al. Evidence for the role of active site residues in the hairpin ribozyme from molecular simulations along the reaction path. J. Am. Chem. Soc. 136, 7789–7792 (2014).
Rupert, P.B., Massey, A.P., Sigurdsson, S.T. & Ferré-D'Amaré, A. Transition state stabilization by a catalytic RNA. Science 298, 1421–1424 (2002).
Desjardins, G., Bonneau, E., Girard, N., Boisbouvier, J. & Legault, P. NMR structure of the A730 loop of the Neurospora VS ribozyme: insights into the formation of the active site. Nucleic Acids Res. 39, 4427–4437 (2011).
Hoffmann, B. et al. NMR structure of the active conformation of the Varkud satellite ribozyme cleavage site. Proc. Natl. Acad. Sci. USA 100, 7003–7008 (2003).
Flinders, J. & Dieckmann, T. A pH controlled conformational switch in the cleavage site of the VS ribozyme substrate RNA. J. Mol. Biol. 308, 665–679 (2001).
Michiels, P.J.A., Schouten, C.H.J., Hilbers, C.W. & Heus, H.A. Structure of ribozyme substrate hairpin of Neurospora VS RNA: A close look at the cleavage site. RNA 6, 1821–1832 (2000).
Cai, Z. & Tinoco, I.J. Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry 35, 6026–6036 (1996).
Butcher, S.E., Allain, F.H. & Feigon, J. Solution structure of the loop B domain from the hairpin ribozyme. Nat. Struct. Biol. 6, 212–216 (1999).
Kraut, J. Serine proteases: structure and mechanism of catalysis. Annu. Rev. Biochem. 46, 331–358 (1977).
Bujnicki, J.M. & Rychlewski, L. Unusual evolutionary history of the tRNA splicing endonuclease EndA: relationship to the LAGLIDADG and PD-(D/E)XK deoxyribonucleases. Protein Sci. 10, 656–660 (2001).
Martick, M. & Scott, W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126, 309–320 (2006).
Wang, S., Karbstein, K., Perecchi, A., Beigelman, L. & Herschlag, D. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry 38, 14363–14378 (1999).
Belousoff, M.J. et al. Ancient machinery embedded in the contemporary ribosome. Biochem. Soc. Trans. 38, 422–427 (2010).
Keel, A.Y., Rambo, R.P., Batey, R.T. & Kieft, J.S. A general strategy to solve the phase problem in RNA crystallography. Structure 15, 761–772 (2007).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Schneider, T.R. & Sheldrick, G.M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Chou, F.-C., Sripakdeevong, P. & Das, R. Correcting pervasive errors in RNA crystallography with Rosetta. Nat. Methods 10, 74–76 (2013).
Pape, T. & Schneider, T.R. HKL2MAP: a graphical user interface for phasing with SHELX programs. J. Appl. Cryst. 37, 843–844 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Keating, K.S. & Pyle, A.M. Semiautomated model building for RNA crystallography using a directed rotameric approach. Proc. Natl. Acad. Sci. USA 107, 8177–8182 (2010).
Wadley, L.M. & Pyle, A.M. The identification of novel RNA structural motifs using COMPADRES: an automated approach to structural discovery. Nucleic Acids Res. 32, 6650–6659 (2004).
Hura, G.L. et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat. Methods 6, 606–612 (2009).
Franke, D., Kikhney, A.G. & Svergun, D.I. Automated acquisition and analysis of small angle X-ray scattering data. Nuc. Inst. Meth. A. 689, 52–59 (2012).
Schneidman-Duhovny, D., Hammel, M., Tainer, J.A. & Sali, A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys. J. 105, 962–974 (2013).
Acknowledgements
We thank J. Kieft for the gift of iridium hexamine; X. Yang, S. Montaño, K. Perry, B. Dhakshnamoorthy and K. Dolan for advice with crystallographic data processing; R. Hulse, I. Dementieva, A. Mateja, F. Qufei, D. Urusova and G. Dobosz for recommendations on growing crystals; and J. Olvera for expression and purification of T7 RNA polymerase. We thank staff of the Advanced Photon Source at Argonne National Laboratory for providing technical advice on X-ray data collection: K. Perry, C. Ogata, N. Venugopalan, B. Nocek and S. Banerjee; and the staff of SIBYLS beam line at Lawrence Berkeley National Laboratory for SAXS data collection. We thank S. Koide, T. Sosnick and S. Crosson for their feedback and discussion on the project, A. Kossiakoff and K. Moffat for use of their data-processing stations, F.-C. Chao and R. Das for their aid with ERRASER software, S. Shelke for help with figures, Y. Shao for help with Amigos II, and N. Tuttle, Q. Dai, N.-S. Li, K. Ceslinski, S. Fica, R. Sengupta, T. Wilson, T. Cech and A. Pyle for discussion and comments on the manuscript. This work was supported by grants from the US National Institutes of Health (R01AI081987 and R01GM102489) to J.A.P. This work is based on research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamline which are supported by a grant from the National Institute of General Medical Sciences (P41 GM103403) from the National Institutes of Health and Advanced Light Source beamline SIBYLS, all supported by US Department of Energy (DOE). This research used resources of the Advanced Photon Source, a US DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract DE-AC02-06CH11357 and Advanced Light Source, a US DOE Office of Science User Facility operated for the DOE Office of Science by Lawrence Berkeley National Laboratory under Integrated Diffraction Analysis (IDAT) grant contract DE-AC02-05CH11231.
Author information
Authors and Affiliations
Contributions
N.B.S., S.D. and J.A.P. designed the study; N.B.S., S.D. and H.H. set up high-throughput crystallization experiments; N.B.S. and S.D. screened crystals and optimized crystallization conditions; N.B.S., S.D. and P.A.R. collected the data; N.B.S. and S.D. phased and solved the structures; N.B.S. collected SAXS data,, J.R.F., S.D. and J.A.P. analyzed the SAXS data. N.B.S., S.D., D.M.J.L., P.A.R. and J.A.P. analyzed the overall data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–4 and Supplementary Figures 1–11. (PDF 2617 kb)
Rights and permissions
About this article
Cite this article
Suslov, N., DasGupta, S., Huang, H. et al. Crystal structure of the Varkud satellite ribozyme. Nat Chem Biol 11, 840–846 (2015). https://doi.org/10.1038/nchembio.1929
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1929
This article is cited by
-
Zn2+-dependent DNAzymes that cleave all combinations of ribonucleotides
Communications Biology (2021)
-
Confluence of theory and experiment reveals the catalytic mechanism of the Varkud satellite ribozyme
Nature Chemistry (2020)
-
Biotechnological and Therapeutic Applications of Natural Nucleic Acid Structural Motifs
Topics in Current Chemistry (2020)
-
High-affinity recognition of specific tRNAs by an mRNA anticodon-binding groove
Nature Structural & Molecular Biology (2019)
-
Big on Change, Small on Innovation: Evolutionary Consequences of RNA Sequence Duplication
Journal of Molecular Evolution (2019)