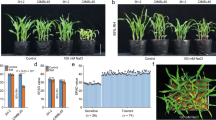
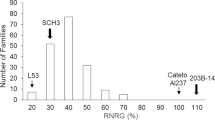
Abstract
The ability of wheat to maintain a low sodium concentration ([Na+]) in leaves correlates with improved growth under saline conditions1,2. This trait, termed Na+ exclusion, contributes to the greater salt tolerance of bread wheat relative to durum wheat3,4. To improve the salt tolerance of durum wheat, we explored natural diversity in shoot Na+ exclusion within ancestral wheat germplasm. Previously, we showed that crossing of Nax2, a gene locus in the wheat relative Triticum monococcum into a commercial durum wheat (Triticum turgidum ssp. durum var. Tamaroi) reduced its leaf [Na+] (ref. 5). Here we show that a gene in the Nax2 locus, TmHKT1;5-A, encodes a Na+-selective transporter located on the plasma membrane of root cells surrounding xylem vessels, which is therefore ideally localized to withdraw Na+ from the xylem and reduce transport of Na+ to leaves. Field trials on saline soils demonstrate that the presence of TmHKT1;5-A significantly reduces leaf [Na+] and increases durum wheat grain yield by 25% compared to near-isogenic lines without the Nax2 locus.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Horie, T., Hauser, F. & Schroeder, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 14, 660–668 (2009).
Munns, R., James, R.A. & Läuchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 57, 1025–1043 (2006).
Dvořák, J., Noaman, M.M., Goyal, S. & Gorham, J. Enhancement of the salt tolerance of Triticum turgidum L. by the Kna1 locus transferred from the Triticum aestivum L. chromosome 4D by homoeologous recombination. Theor. Appl. Genet. 87, 872–877 (1994).
Gorham, J., Wyn Jones, R.G. & Bristol, A. Partial characterisation of the trait for enhanced K+-Na+ discrimination in the D genome of wheat. Planta 180, 590–597 (1990).
James, R.A., Davenport, R.J. & Munns, R. Physiological characterisation of two genes for Na+ exclusion in durum wheat: Nax1 and Nax2. Plant Physiol. 142, 1537–1547 (2006).
Rengasamy, P. Soil processes affecting crop production in salt-affected soils. Funct. Plant Biol. 37, 613–620 (2010).
Tilman, D., Balzer, C., Hill, J. & Belfort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 108, 20260–20264 (2011).
Munns, R. & Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681 (2008).
Dubcovsky, J., María, G.S., Epstein, E., Luo, M.C. & Dvořák, J. Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor. Appl. Genet. 92, 448–454 (1996).
Huang, S., Spielmeyer, W., Lagudah, E.S. & Munns, R. Comparative mapping of HKT genes in wheat, barley and rice, key determinants of Na+ transport and salt tolerance. J. Exp. Bot. 59, 927–937 (2008).
Davenport, R.J., James, R.A., Zakrisson-Plogander, A., Tester, M. & Munns, R. Control of sodium transport in durum wheat. Plant Physiol. 137, 807–818 (2005).
Byrt, C.S. et al. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 143, 1918–1928 (2007).
Schachtman, D.P. & Schroeder, J.I. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370, 655–658 (1994).
Maser, P. et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 531, 157–161 (2002).
Davenport, R.J. et al. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 30, 497–507 (2007).
Møller, I.S. et al. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21, 2163–2178 (2009).
Ren, Z.H. et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37, 1141–1146 (2005).
Plett, D. et al. Improved salinity tolerance of rice through cell type-specific expression of AtHKT1;1. PLoS ONE 5, e12571 (2010).
Sunarpi et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 44, 928–938 (2005).
Jabnoune, M. et al. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. 150, 1955–1971 (2009).
Läuchli, A., James, R.A., Munns, R., Huang, C. & McCully, M. Cell-specific localization of Na+ in roots of durum wheat and possible control points for salt exclusion. Plant Cell Environ. 31, 1565–1574 (2008).
Gassmann, W., Rubio, F. & Schroeder, J.I. Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. Plant J. 10, 869–882 (1996).
Yao, X. et al. Differential sodium and potassium transport selectivities of the rice OsHKT2;1 and OsHKT2;2 transporters in plant cells. Plant Physiol. 152, 341–355 (2010).
Uozumi, N. et al. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 122, 1249–1260 (2000).
Yeo, A.R. & Flowers, T.J. Salinity resistance in rice (Oryza sativa L.) and a pyramiding approach to breeding varieties for saline soils. Aust. J. Plant Physiol. 13, 161–173 (1986).
Schachtman, D.P., Munns, R. & Whitecross, M.I. Variation in sodium exclusion and salt tolerance in Triticum tauschii. Crop Sci. 31, 992–997 (1991).
Munns, R. & James, R.A. Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253, 201–218 (2003).
James, R.A. et al. Photosynthetic capacity is related to the cellular and subcellular partitioning of Na+, K+ and Cl− in salt-affected barley and durum wheat. Plant Cell Environ. 29, 2185–2197 (2006).
Richards, R.A. Should selection for yield in saline regions be made on saline or non-saline soils? Euphytica 32, 431–438 (1983).
James, R.A., Blake, C., Byrt, C.S. & Munns, R. Major genes for Na+ exclusion Nax1 and Nax2 (wheat HKT1;4 and HKT1;5) decrease Na+ accumulation in bread wheat under saline and waterlogged conditions. J. Exp. Bot. 62, 2939–2947 (2011).
Rodríguez-Navarro, A. & Ramos, J. Dual system for potassium transport in Saccharomyces cerevisiae. J. Bacteriol. 159, 940–945 (1984).
Molendijk, A.J. et al. A cysteine-rich receptor-like kinase NCRK and a pathogen-induced protein kinase RBK1 are Rop GTPase interactors. Plant J. 53, 909–923 (2008).
Gietz, R.D. & Schiestl, R.H. High efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 (2007).
Roy, S.J. et al. Investigating glutamate receptor-like gene co-expression in Arabidopsis thaliana. Plant Cell Environ. 31, 861–871 (2008).
Dick, D.A.T. & McLaughlin, S.G. The activities and concentration of sodium and potassium in toad oocytes. J. Physiol. (Lond.) 205, 61–78 (1969).
Conn, S.J. et al. Magnesium transporters, MGT2/MRS2-1 and MGT3/MRS2-5, are important for magnesium partitioning within Arabidopsis thaliana mesophyll vacuoles. New Phytol. 190, 583–594 (2011).
Gilliham, M., Athman, A., Tyerman, S.D. & Conn, S.J. Cell-specific compartmentation of mineral nutrients is an essential mechanism for optimal plant productivity; another role for TPC1? Plant Signal. Behav. 6, 1656–1661 (2011).
Koltai, H. & Bird, D.M. High throughput cellular localization of specific plant mRNAs by liquid-phase in situ reverse transcription-polymerase chain reaction of tissue sections. Plant Physiol. 123, 1203–1212 (2000).
Acknowledgements
The authors thank grain growers A. and J. Crowe for hosting our field trials on their property at Moree. We acknowledge the assistance of C. Blake (CSIRO), B. Collard, D. Gulliford (NSW DPI) and A. Smart (PCT); J. Dubcovsky, University of California, Davis, for kindly providing the BAC sequence used to design cslinkNax2; D. Cooper, T. Rathjen, A. Webster and J. Able for additional field data on Nax2. We thank S. Ramesh for extracting Xenopus oocytes, Adelaide Microscopy for facilities and training, G. Mayo for assistance with in situ PCR, S. Roy for assistance with confocal microscopy and flame photometry and O. Cotsaftis for the clone of OsHKT1;5. This research was supported by the Grains Research and Development Corporation (GRDC) of Australia and the Australian Research Council (ARC).
Author information
Authors and Affiliations
Contributions
R.M., R.A.J., R.A.H., M.T., D.P. and M.G. conceived the project and planned experiments. R.M. and M.G. supervised the research. B.X. performed all Xenopus, yeast and protoplast experiments and R.A.J. performed field research. C.S.B. performed wheat genotyping. S.D.T. assisted with electrophysiology experiments. S.J.C., A.A. and C.J. performed in situ PCR and qPCR. M.G., D.P., R.A.J. and R.M. wrote the manuscript. All authors commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–4 and Supplementary Figs. 1–4 (PDF 486 kb)
Rights and permissions
About this article
Cite this article
Munns, R., James, R., Xu, B. et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol 30, 360–364 (2012). https://doi.org/10.1038/nbt.2120
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2120