Abstract
Pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) in plants enable them to respond to pathogens by activating the production of defence metabolites that orchestrate immune responses1,2,3,4. How the production of defence metabolites is promoted by immune receptors and coordinated with broad-spectrum resistance remains elusive. Here we identify the deubiquitinase PICI1 as an immunity hub for PTI and ETI in rice (Oryza sativa). PICI1 deubiquitinates and stabilizes methionine synthetases to activate methionine-mediated immunity principally through biosynthesis of the phytohormone ethylene. PICI1 is targeted for degradation by blast fungal effectors, including AvrPi9, to dampen PTI. Nucleotide-binding domain, leucine-rich-repeat-containing receptors (NLRs) in the plant immune system, such as PigmR, protect PICI1 from effector-mediated degradation to reboot the methionine–ethylene cascade. Natural variation in the PICI1 gene contributes to divergence in basal blast resistance between the rice subspecies indica and japonica. Thus, NLRs govern an arms race with effectors, using a competitive mode that hinges on a critical defence metabolic pathway to synchronize PTI with ETI and ensure broad-spectrum resistance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data are available within this Article and its Supplementary Information. Original gel blots are shown in Supplementary Fig. 1. Original data points in graphs are shown in the Source Data files. Statistical analyses of this study are provided in Supplementary Table 4. The sequences of PICI1, OsMETS1 and OsMETS2 have been deposited and made publicly available in GenBank with accession codes MT920667, MT920668 and MT920669, respectively. Protein structure models of Protein Data Bank (PDB) ID 2WP7 and PDB ID 1U1U were obtained from the PDB. Source data are provided with this paper.
References
Jones, J. D. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Couto, D. & Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552 (2016).
Xin, X.-F. Kvitko, B. & He, S. Y. Pseudomonas syringae: what it takes to be a pathogen. Nat. Rev. Microbiol. 16, 316–328 (2018).
Kourelis, J. & van der Hoorn, R. A. L. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30, 285–299 (2018).
Li,W., Deng, Y., Ning, Y., He, Z. & Wang, G. L. Exploiting broad-spectrum disease resistance in crops: from molecular dissection to breeding. Annu. Rev. Plant Biol. 71, 575–603 (2020).
Ngou, B. P. M., Ahn, H.-K., Ding, P. & Jones, J. D. G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115 (2021).
Yuan, M. et al. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109 (2021).
Bürger, M. & Chory, J. Stressed out about hormones: how plants orchestrate immunity. Cell Host Microbe 26, 163–172 (2019).
Broekaert, W. F., Delauré Sl, M. F. C., Bolle, D., De Bolle, Mf, B. P. A. & Cammue, B. P. The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 44, 393–416 (2006).
Washington, E. J. et al. Pseudomonas syringae type III effector HopAF1 suppresses plant immunity by targeting methionine recycling to block ethylene induction. Proc. Natl Acad. Sci. USA 113, E3577–E3586 (2016).
Zhao, S. et al. A viral protein promotes host SAMS1 activity and ethylene production for the benefit of virus infection. eLife 6, e27529 (2017).
Dean, R. et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430 (2012).
Mosquera, G., Giraldo, M. C., Khang, C. H., Coughlan, S. & Valent, B. Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1–4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell 21, 1273–1290 (2009).
Mentlak, T. A. et al. Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 24, 322–335 (2012).
Orbach, M. J., Farrall, L., Sweigard, J. A., Chumley, F. G. & Valent, B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12, 2019–2032 (2000).
Kanzaki, H. et al. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 72, 894–907 (2012).
Sweigard, J. A. et al. Identification, cloning, and characterization of PWL2, a gene for host species-specificity in the rice balst fungus. Plant Cell 7, 1221–1233 (1995).
Wu, J. et al. Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice. New Phytol. 206, 1463–1475 (2015).
Yoshida, K. et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21, 1573–1591 (2009).
Li, W. et al. The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant Microbe Interact. 22, 411–420 (2009).
Deng, Y. et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965 (2017).
Xie, Z. et al. A nucleotide-binding site-leucine-rich repeat receptor pair confers broad-spectrum disease resistance through physical association in rice. Phil. Trans. R. Soc. B 374, 20180308 (2019).
Zhou, B. et al. The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol. Plant Microbe Interact. 19, 1216–1228 (2006).
Xie, X. et al. PPPDE1 promotes hepatocellular carcinoma development by negatively regulate p53 and apoptosis. Apoptosis 24, 135–144 (2019).
Shimizu, T. et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214 (2010).
Suh, H. Y. et al. Crystal structure of DeSI-1, a novel deSUMOylase belonging to a putative isopeptidase superfamily. Proteins 80, 2099–2104 (2012).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Chen, X. L. et al. Proteomic analysis of ubiquitinated proteins in rice (Oryza sativa) after treatment with pathogen-associated molecular pattern (PAMP) elicitors. Front. Plant Sci. 9, 1064 (2018).
Iwai, T., Miyasaka, A., Seo, S. & Ohashi, Y. Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol. 142, 1202–1215 (2006).
Singh, R. et al. Magnaporthe oryzae effector AVR-Pii helps to establish compatibility by inhibition of the rice NADP-malic enzyme resulting in disruption of oxidative burst and host innate immunity. Mol. Cells 39, 426–438 (2016).
De la Concepcion, J. C. et al. Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen. Nat. Plants 4, 576–585 (2018).
Maqbool, A. et al. Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife 4, 08709 (2015).
Wang, W. et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49 (2018).
Huang, X. et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44, 32–39 (2011).
Hu, C. et al. Metabolic variation between japonica and indica rice cultivars as revealed by non-targeted metabolomics. Sci. Rep. 4, 5067 (2014).
Liao, J. et al. Pathogen effectors and plant immunity determine specialization of the blast fungus to rice subspecies. eLife 5, e19377 (2016).
Axtell, M. J. & Staskawicz, B. J. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377 (2003).
Mackey, D., Holt, B. F. III, Wiig, A. & Dangl, J. L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754 (2002).
He, J. et al. A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol. Plant 12, 1561–1576 (2019).
Ma, X. et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot Plants. Mol. Plant 8, 1274–1284 (2015).
Zhai, K. et al. RRM transcription factors interact with NLRs and regulate broad-spectrum blast resistance in rice. Mol. Cell 74, 996–1009 (2019).
Park, C.-H. et al. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24, 4748–4762 (2012).
Acknowledgements
We thank J. Zhou and J. Li for critical reading and discussion; X. Wei and G. Zhang for providing the rice germplasm and CSSL; J. Zhang for help with ethylene measurement; S. Wang for help with Met measurement; B. Liu, C. Wang and Z. Gu for help with bioinformatic analysis; and X. Fu for sharing HA-Ub plasmids. This work was supported by grants from the National Natural Science Foundation of China (32088102, 31720103913 and U20A2021), the Chinese Academy of Sciences (XDB27040201 and XDA24010304), the National Key Research and Development Program of China (2016YFD0100600), the Science and Technology Commission of Shanghai Municipality (19391900300), the Project of Special Funding for Crop Science Discipline Development of Yangzhou University (yzuxk202006) and the National Key Laboratory of Plant Molecular Genetics. K.Z. was supported by the National Postdoctoral Program for Innovative Talents (BX20190346) and by the Shanghai Postdoctoral Excellence Program.
Author information
Authors and Affiliations
Contributions
K.Z. and Z.H. conceived and designed the experiments. K.Z. and D.L. performed most experiments, including the Y2H, SLC, BiFC, co-IP, subcellular localization, cell-free degradation assay, deubiquitination assay, ROS detection and western blots. K.Z., H.L. and Y.W. performed Met and ethylene measurement. K.Z., H.L. and F.J. performed RNA analysis. K.Z., D.L., B.Y. and X.G. performed pathoassays. D.L. and H.L. performed agronomic traits analysis. K.Z., J.L., Z.L., L.H., X.W. and J.-Y.L. generated material used in this study. J.M. performed the domestication analysis. H.G. and B.H. supervised Y.W. and J.M., respectively. L.Z., E.W., Y.D., C.-K.W., H.G. and B.H. provided theoretical contributions to the project. K.Z., D.L. and Z.H. analysed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Nicholas Talbot and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
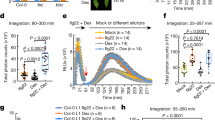
Extended Data Fig. 1 Screening of PICIs and PigmR–PICI1 interaction analysis.
a, The workflow for identifying the proteins involved in both PTI and ETI. Tandem Mass Tag (TMT)-based proteomic analysis was performed to investigate the global changes of rice protein profiling with chitin treatment, the blue circle denotes the chitin-induced 413 candidate proteins. Total 11 PigmR-CC interacting proteins were identified through yeast-two-hybrid screen and shown with the pink circle. The overlap between the blue and pink circles indicates 3 PICI candidates. b, Protein alignment of PICI1 and its homologs in A. thaliana, S. cerevisiae, C. elegans, M. musculus and H. sapiens (NP_194296.1, NP_594707.2, NP_741592.1, NP_077244.1 and NP_057160.2, respectively) with MegAlign software. The conserved PPPDE region is underlined in red and the conserved site Cys115 residue is marked with red arrow. c, Immunodetection of protein expression in yeast. pDEST22 (EV), pDEST22-PICI1 (1) and pDEST22-Os07g16130 (2) detected using anti-GAL4-AD antibody. pDEST32-PigmR (BD-PigmR), pDEST32-PigmR-CC (BD-PigmR-CC), pDEST32-PigmR-NBS (BD-NBS) and pDEST32-PigmR-LRR (BD-LRR) detected using anti-GAL4-BD antibody. The asterisks indicate the target proteins. d, Protein alignment of PICI1 and its homologs in rice. e, PigmR and PigmR-CC specifically interact with PICI1 but not its homologs in Y2H assay. f, Immunodetection of protein levels in yeast. Upper panel (AD), pDEST22 (lane 1), pDEST22-PICI1 (lane 2), -Os02g43840 (lane 3), -Os03g01130 (lane 4), -Os03g10200 (lane 5), -Os04g46290 (lane 6), -Os06g01780 (lane 7), -Os06g08360 (lane 8), -Os06g36490 (lane 9), detected using anti-GAL4-AD antibody; middle panel (BD), pDEST32-PigmR-CC, detected using anti-GAL4-BD antibody. The asterisks indicate the target proteins. g, SLC assay of PigmR-PICI1 interaction in N. benthamiana. PIBP1 and Os07g16130 served as positive and negative control, respectively. Fluorescence signal intensity is indicated. h, Western blot confirming expression of proteins in N. benthamiana. Note that cLuc-PICI1 was detected using anti-PICI1 antibody. Short, long = short or long exposure. i, PICI1-YFP was co-expressed with various known organelle markers as indicated in rice protoplasts. mCherry = cytoplasmic marker, NLS-RFP = nucleus marker, ST-mCherrry = trans-Golgi cisternae marker, SYP61-mCherry = trans-Golgi network/early endosome (TGN/EE) marker, ER-mCherry = endoplasmic reticulum marker, OsRac1-mCherrry = cell member marker, Chl Auto = Chloroplast autofluorescence. j, Western blot confirming expression of PICI1-YFP without free YFP in rice protoplast. k, Localization of PICI1-GFP in representative root cells of stable transgenic plants. DAPI staining indicates the nucleus. l, Bimolecular fluorescence complementation (BiFC) verifies the PigmR-CC/PICI1 interaction in rice protoplasts. Pish-CC served as a negative control. Ponceau S staining (c, f) or Actin (h, j) served as loading control. Scale bars, 5 µm (i, l) or 10 μm (k). Experiments were repeated twice (c, f, h) or three times with similar results (e, g, i–l).
Extended Data Fig. 2 PICI1 is involved in PigmR-, Pizt- and Pi9- but not Pish-mediated blast resistance.
a, i, l, o, Schematic of two independent PICI1 knockout lines in NIL-Pigm (a), ZH11 (Pizt-containing) (i), Ky-Pi9 (Pi9-containing) (l) and NIPB (Pish-containing) (o), respectively. b, j, m, p, Protein analysis of PICI1 in the corresponding transgenic plants using an anti-PICI1 antibody. The asterisk indicates the endogenous PICI1 and the band around 63 kDa indicates the PICI1-GFP fusion protein (b, p). c, Blast resistance of NIL-Pigm, PICI1-KO/NIL-Pigm and PICI1-OE/NIL-Pigm lines at 7dpi with punch injection inoculation (TH12). NIPB served as a susceptible control. d, Y2H assay of interactions between PICI1 and the CC domains of PigmR, Pi9, Pizt and Pish. EV, empty vector. e, Immunodetection of protein levels in yeast. Upper panel (AD), pDEST22-PICI1, detected using anti-GAL4-AD antibody; middle panel (BD), PigmR-CC, Pi9-CC, Pizt-CC and Pish-CC, detected using anti-GAL4-BD antibody. f, SLC confirmation of the PICI1-Pizt-CC and PICI1/Pi9-CC interactions in N. benthamiana. PigmR-CC and Pish-CC served as positive and negative control, respectively. Fluorescence signal intensity is indicated. g, Relative luciferase activity of protein-protein interactions was measured. One-way ANOVA with Tukey’s test (mean ± s.d.; n = 5, biologically independent samples). Different letters indicate significant difference at P < 0.05. h, Western blot analysis confirming the expression level of each protein in N. benthamiana. k, n, q, Punch inoculation of PICI1-KO transgenic plants in ZH11 (k), Ky-Pi9 (n) and NIPB (q). Blast resistance of WT and two representative transgenic lines, 5 dpi with avirulent strain YN2 (k, q) or Guy11 (n). TH12 served as a susceptible control (q). r, Transient assay of cell death in rice protoplast. Relative luciferase activity was measured after transformation with avrPizt-mCherry in rice protoplasts derived from ZH11 or PICI1-KO/ZH11. Empty vector (EV) was used as a negative control. Two-tailed Student’s t-test (mean ± s.d.; n = 9, biologically independent samples). s, Western blot analysis confirming expression of proteins in rice protoplasts. Actin (b, h, j, m, p, s) or Ponceau S staining (e) served as loading control. The asterisks indicate the target proteins. For c, k, n, q, data were analysed by two-tailed Student’s t-test. Fungal growth (mean ± s.d.; n = 3, biologically independent samples), lesion lengths or areas (mean ± s.d.; n = numbers of biologically independent samples in the graphs). Scale bars, 1 cm. Asterisks represent significant difference (*P < 0.05, **P < 0.01) (c, k, n, q, r). Exact P values are provided in Supplementary Table 4 (c, g, k, n, q, r). Experiments were repeated twice (b, e, h, j, m, p) or three times with similar results (c, d, f, g, k, n, q, r, s).
Extended Data Fig. 3 PICI1 is involved in chitin- and flg22-induced PTI responses.
a, g, p, Induction of the PTI-related defence genes OsPAL1 and KS4 at 1 h after chitin (a, g) or flg22 (p) incubation in WT, PICI1-KO and PICI1-OE lines in NIL-Pigm (a, p) or NIPB (g) background. Two-tailed Student’s t-test (mean ± s.d.; n = 3, biologically independent samples). b, c, Induction of PICI1 in NIL-Pigm upon chitin/flg22 treatment (b) and during M. oryzaeTM21 infection (c) with water as control. d, h, PICI1 was induced upon M. oryzae TM21 infection (d) and chitin treatment (h) with water as control. The asterisk indicates endogenous PICI1 (d). The relative PICI1 protein abundance was indicated. e, Induction of PTI-related defence genes KS4 and OsPAL1 in NIL-Pigm leaves during M. oryzae TM21 infection. f, o, Chitin- (f) or flg22- (o) induced ROS burst in WT, PICI1-KO and PICI1-OE lines in NIPB (f) or NIL-Pigm (o) background. Data are mean ± s.d. (n = 9, biologically independent samples). i, Disease resistance of PICI1 transgenic plants in NIPB. j, l, Schematic of two independent PICI1/OsCERK1 double knockout (j) and PICI1-OE/OsCERK1-KO (l) lines in NIPB. k, m, Protein levels of PICI1 (k) or PICI1-GFP (m) in PICI1/OsCERK1-KO (k) or PICI1-OE/OsCERK1-KO (m). n, Punch inoculation of OsCERK1-KO, PICI1/OsCERK1-KO and PICI1-OE/OsCERK1-KO transgenic lines. q, r, PICI1 was induced upon flg22 treatment (q) and bacterial Xoo (strain, PXO99A) infection (r) with water as control. s–u, Disease resistance to bacterial Xoo in the PICI1 transgenic lines in NIL-Pigm. Triangles represent the end of bacterial infection sites. For i, n, t, u, data were analysed by two-tailed Student’s t-test. Lesion lengths (mean ± s.d.; n = numbers of biologically independent samples in the graphs), fungal or bacterial growth (mean ± s.d.; n = 3, biologically independent samples). For b, c, e, data are mean ± s.d. (n = 3, biologically independent samples). The rice ACTIN1 served as an internal control (a-c, e, g, p). Actin was detected as a loading control (d, h, k, m, q, r). Blast resistance of representative transgenic lines, 5 dpi with virulent strain TH12 (i, n). Scale bars, 1 cm (i, n) or 5 cm (s). Asterisks represent significant difference (*P < 0.05, **P < 0.01) (a, g, i, n, p, t, u). Experiments were repeated twice (k, m) or three times with similar results (a–i, n, o–u).
Extended Data Fig. 4 PICI1 functions as a deubiquitinase, and interacts with and stabilizes OsMETS1.
a, His-PICI1 and His-PICI1-PPPDE displayed cleavage activity toward K48- and K63-linked but not linear ubiquitin in vitro. His alone served as a control. b, Homology model of PICI1-PPPDE domain (cyan) aligned with Desi-1 (PDB ID code 2WP7) (blue) using PyMOL. The putative catalytic dyad residues of H41 and C115 in PICI1 are shown in yellow and the catalytic residues of Desi-1 H38 and C108 are shown in magenta. The boxed section is magnified for close view. c, Induction of KS4, PR10 and PR5 in PICI1-YFP or PICI1C115S-YFP overexpression protoplast after chitin treatment (1 hpi) with the empty vector control. Lowercase letters indicate statistical significance (P < 0.05). One-way ANOVA with Tukey’s test. Data are mean ± s.d. (n = 3, biologically independent samples). Exact P values are provided in Supplementary Table 4. d, Ubiquitin conjugation in total protein extracts from 2-week-old NIL-Pigm and PICI1-KO/NIL-Pigm, after spraying TH12 or TM21 with water as control for 36 h. e, Morphological phenotype of mature plants of wild-type and PICI1-KO/NIL-Pigm. No obvious change in morphology was observed in the PICI1-KO plants. Scale bar, 10 cm. f, Venn diagram showing the number of total ubiquitinated proteins (blue), PICI1 interacting candidates identified in IP–MS (red), PTI-related ubiquitinated proteins (green) and overlapping proteins. g, Protein alignment of OsMETS1 and its homologs in sorghum, soybean, maize, wheat and Arabidopsis (XP_021301657.1, XP_003542326.1, PWZ52049.1, TraesCS4D02G012900.2 and XP_002871787.1) with MegAlign software. h, Ethylene biosynthesis pathway in plants. S-AdoMet, S-adenosylmethionine; Hcys, homocysteine; THF, tetrahydrofolate; SAH, S-adenosylhomocysteine; Ado, adenosine; ACS, 1-aminocyclopropane-1-carboxylate synthase; ACC, 1-aminocyclopropane-1-carboxylate. i, A SLC assay of OsMETS1 and PICI1 interaction in N. benthamiana. PICI2 served as a negative control. Fluorescence signal intensity is indicated. j, Western blot confirming protein expression in N. benthamiana. k, GST-PICI1 pulls down His-OsMETS1 in vitro. l, Subcellular localization of OsMETS1-GFP in OsMETS1-GFP/NIL-Pigm root. DAPI staining indicates the nucleus. Scale bars, 10 μm. m, Immunodetection of OsMETS1-GFP in transgenic lines, with NIL-Pigm as a negative control. n, Subcellular localization of OsMETS1 and co-localization of PICI1-YFP/OsMETS1-mCherry in rice protoplasts. Scale bars, 5 µm. o, Western blot confirming expression of OsMETS1-mCherry without free mCherry in rice protoplast. mCherry (lane 1), OsMETS1-mCherry (lane 2). p, Western blot confirming expression of OsMETS1-YFP without free YFP in rice protoplast. YFP (lane 1), OsMETS1-YFP (lane 2). q, Degradation of OsMETS1-Flag was proteasome-dependent and delayed by PICI1 in cell-free system. r, PICI1 stabilizes OsMETS1-Flag in rice plants. The lysates from WT, PICI1-KO and PICI1-OE plants were co-incubated with OsMETS1-Flag in the presence or absence of MG132. s, Peptide coverage and ubiquitination sites identified in OsMETS1 by tandem mass spectrometry after M. oryzae (TH12) or water treatment for 36 h. The various peptides identified by mass spectrometry are shown in blue; the lysine acceptor sites detected in red are numbered below with positions; the four specific deubiquitination sites are denoted with asterisks. t, Structure model for OsMETS1. Homology model of the OsMETS1 based on the crystal structure of Arabidopsis Met synthase (PDB ID code 1U1U) and analysed by PyMOL. The four lysines identified as deubiquitination sites were shown in blue. The other lysine acceptor sites were shown in red. u, Cell-free degradation assays show the proteasome-dependent degradation of His-OsMETS1 and delayed degradation of His-OsMETS1 mutant variants. Purified recombinant His-OsMETS1 and its mutants were incubated with protein extracts from non-infected NIPB. His alone served as a negative control. v, The OsMETS1 mutants showed higher protein accumulation compared to wild-type OsMETS1 in protoplasts. The YFP tag protein was expressed as an internal control. The transcript levels were determined by semi-quantitative PCR. Actin was detected as a loading control (d, j, m, o, p). Ponceau S staining served as loading control (q, r, u). The protein abundance was quantified using ImageJ and indicated under lanes (q, r, u). Experiments were repeated twice (j, m, o, p) or three times with similar results (a, c, d, i, k, l, n, q, r, u, v).
Extended Data Fig. 5 PICI1 directly deubiquitinates and stabilizes OsMETS.
a, Induction of PR5 and PR1 in OsMETS1, OsMETS1K28R, OsMETS1K196R, OsMETS1K458R and OsMETS1K740R overexpression protoplasts after chitin treatment (1 hpi). Lowercase letters indicate statistical significance (P < 0.05). One-way ANOVA with Tukey’s test. Data are mean ± s.d. (n = 3, biologically independent samples). Exact P values are provided in Supplementary Table 4. b, Cell-free degradation assays show the delayed degradation of His-OsMETS14KR mutant variant, compared to His-OsMETS1K28R. Purified recombinant fusion proteins were incubated with non-infected NIPB extracts. c, The OsMETS14KR mutant showed higher protein accumulation compared to wild-type OsMETS1 and OsMETS1K28R in protoplasts. The YFP tag protein was expressed as an internal control. The transcript levels were determined by semi-quantitative PCR. d, Stability of the OsMETS1-Flag depends on His-PICI1 but not the His-PICI1C115S in cell-free degradation assay. e, Deubiquitination analysis of ubiquitin-modified OsMETS1-GFP, immunoprecipitated from OsMETS1-GFP/NIL-Pigm plants, by incubating with recombinant His-PICI1 or His-PICI1C115S respectively. f, Tail-to-head organization of OsMETS1 and OsMETS2 genes in rice. g, Protein alignment of OsMETS1 and OsMETS2 with MegAlign software. h, Co-IP assay of PICI1 with OsMETS2 in rice. PICI2 served as a negative control. i, A SLC assay showed that both OsMETS1 and OsMETS2 interacted with PICI1 in N. benthamiana. PICI2 was used as a negative control. Fluorescence signal intensity is indicated. j, Western blot confirming protein expression in N. benthamiana. k, Cell-free degradation assay showed the proteasome-dependent degradation of OsMETS2-Flag in N. benthamiana. l, Cell-free degradation assay showed the delayed degradation of OsMETS2 in presence of recombinant His-PICI1. m, Co-IP assay of PICI1 with endogenous OsMETS in rice. The GFP negative control and PICI1-GFP were constitutively expressed in transgenic rice plants. anti-OsMETS, recognizing both OsMETS1 and OsMETS2. n, Cell-free degradation assay showed that endogenous OsMETS degradation was delayed by PICI1. o, Immunodetection of the recombinant His-OsMETS1 and His-OsMETS2 using the anti-OsMETS antibody. His alone served as a negative control. p, A schematic diagram of two independent OsMETS1-KO lines in NIL-Pigm background. q, Protein levels of OsMETS in NIL-Pigm, OsMETS1-GFP/NIL-Pigm and OsMETS1-KO/NIL-Pigm plants using the anti-OsMETS antibody. The band around 75 kDa indicates the endogenous OsMETS. r, Relative transcript levels of OsMETS in the wild-type, PICI1-KO and PICI1-OE lines in NIL-Pigm and NIPB background. Data are mean ± s.d. (n = 3, biologically independent samples). s, Analysis of endogenous OsMETS ubiquitination. OsMETS was immunoprecipitated from wild-type NIL-Pigm with anti-OsMETS or anti-IgG as negative control. Short, long = Short or long exposure. The protein abundance was quantified using ImageJ and indicated under lanes (b, d, k, l, n, q). Ponceau S staining (b, d, k, l, n, o) or Actin (e, j, q, s) was used as a control for equal loading. The rice ACTIN1 served as an internal control (a, r). Experiments were repeated twice (h, j, m, n, o, s) or three times (a–e, i, k, l, q, r) with similar results.
Extended Data Fig. 6 OsMETS functions in rice PTI and ETI.
a, b, c, d, OsMETS is induced upon treatment of chitin (a) or flg22 (b), and infection of Xoo (PXO99A) (c) or M. oryzae (TM21) (d). e, Protein accumulation of OsMETS in NIL-Pigm, PICI1-KO/NIL-Pigm and PICI1-GFP/NIL-Pigm in early infection stage (TM21). f, g, Decreased ubiquitination of OsMETS1-GFP in PTI (f) and ETI (g). The protein extracts from OsMETS1-GFP/NIL-Pigm plants incubated with chitin for 1h (f) or infected with TH12 for 36 h (g), with water as control, was analysed. NIL-Pigm served as a negative control. h, A schematic diagram of two independent OsMETS2-KO lines in NIL-Pigm. i, Decreased protein levels of OsMETS in OsMETS2-KO/NIL-Pigm compared with NIL-Pigm. j, l, Basal blast resistance of NIL-Pigm, OsMETS2-KO/NIL-Pigm (j) and OsMETS2-GFP/NIL-Pigm (l) at 5 dpi with punch inoculation (TM21). k, Immunodetection of the OsMETS2-GFP in the two independent transgenic lines, with NIL-Pigm as a negative control. m, n, Disease resistance of WT, OsMETS1-KO (m), OsMETS1-OE (m) and OsMETS2-KO (n) were shown, 5 dpi with TH12, NIPB served as a susceptible control. o, Relative transcript levels of OsMETS1, OsMETS2 and OsMETS in NIL-Pigm and two OsMETS-RNAi lines. p, Protein levels of OsMETS in NIL-Pigm and OsMETS-RNAi lines. q, Morphological phenotype of NIL-Pigm and OsMETS-RNAi/NIL-Pigm lines at mature stage in the paddy field. r, OsMETS-RNAi/NIL-Pigm plants significantly reduced plant height (upper panel), tiller number (middle panel), grain yield per plant (lower panel). s, Relative Met contents in WT and OsMETS-RNAi/NIL-Pigm. t, Increased Met accumulation in NIL-Pigm seedling shoots after root incubation with 25 mM Met for 48 h. u, Increased blast resistance after Met treatment in NIPB. v, Induction of the defence gene KS4 at 48 h after root dipping with 25 mM Met solution or water in NIL-Pigm. w, y, Basal blast resistance of Met-treated OsMETS1-KO/NIL-Pigm at 5 dpi (w) or PICI1-KO/NIL-Pigm at 7 dpi (y). Met solution (10 mM) or water was supplied from the roots for 48 h and plants were then inoculated with TM21. x, Relative Met contents in WT and PICI1-KO/NIL-Pigm lines. For j, l, m, n, data were analysed by two-tailed Student’s t-test. Lesion lengths (mean ± s.d.; n = numbers of biologically independent samples in the graphs), fungal growth (mean ± s.d.; n = 3, biologically independent samples). For u, w, y, one-way ANOVA with Tukey’s test. Lesion areas (mean ± s.d.; n = numbers of biologically independent samples in the graphs), fungal growth (mean ± s.d.; n = 3, biologically independent samples). Lowercase letters indicate statistical significance (P < 0.05). For r, s, t, x, data were analysed by two-tailed Student’s t-test (mean ± s.d.; n = numbers of biologically independent samples in the graphs). In o, v, two-tailed Student’s t-test (mean ± s.d.; n = 3, biologically independent samples). Asterisks indicate significant differences (*P < 0.05, **P < 0.01) (j, l-o, r–t, v, x). Exact P values are provided in Supplementary Table 4 (j, l–o, r–y). Scale bars, 1 cm (j, l–n, u, w, y), 10 cm (q). Relative protein abundance was indicated below (a–e, p). Actin was detected as a loading control (a–g, i, k, p). The rice ACTIN1 served as an internal control (o, v). Experiments were independently repeated twice (i, k, p, r) or three times (a–g, j, l–o, s–y) with similar results.
Extended Data Fig. 7 PICI1–OsMETS module confers blast resistance through regulating Met–ethylene biosynthesis.
a, High Met concentrations (10 and 25 mM) inhibit blast fungal growth on medium. The M. oryzae isolate TH12 was inoculated on the complete medium (CM) with Met supplement. The colony diameters were measured. One-way ANOVA with Tukey’s test (mean ± s.d.; n = 5, biologically independent samples). b, c, Ethylene contents in WT and transgenic lines of OsMETS1 (OsMETS1-KO and OsMETS1-OE) (b) and PICI1 (PICI1-KO and PICI1-OE) (c) infected by TM21 (2-week-old, 36 hpi). Two-tailed Student’s t-test (mean ± s.d.; n = 3, biologically independent experiments). d, Induction of ERF63 and ERF73 by ethylene treatment in NIL-Pigm. Two-week-old seedlings were placed into sealed containers with 10 or 100 ppm (μl/L) ethylene, or air as control for 3 h to analyse gene expression. e, f, Suppression of OsMETS1 (e) and OsMETS2 (f) expression by ethylene treatment (100 ppm) in NIL-Pigm. g, Relative expression of ERF63 at 1 d treatment with 10 μM AVG or 20 μM ACC or water. ERF63 was chosen as a marker gene for AVG and ACC responses. Two-tailed Student’s t-test (mean ± s.d.; n = 3, biologically independent samples). h, Effects of AVG and ACC on basal blast resistance in NIPB. Two-week-old seedlings were pre-treated with 10 μM AVG, or 20 μM ACC or water for 1 d and then spray-inoculated with rice blast (TH12), 5 dpi. i, j, Basal blast resistance of ACC-treated PICI1-KO/NIL-Pigm (i) and OsMETS-RNAi/NIL-Pigm (j) plants. ACC solution (20 μM) or water was supplied from the leaves and inoculated with TM21, 5 dpi. k, l, Induction of OsACS2 in NIL-Pigm leaves at early stage by TM21 (k, PTI) or late stage by TH12 (l, ETI) infection. m, Enhanced ethylene production in NIL-Pigm after rice blast infection (TH12 or TM21), with water as a control for 36 h. One-way ANOVA with Tukey’s test (mean ± s.d.; n = 3, biologically independent experiments). n, A schematic diagram of OsACS2-KO lines in NIL-Pigm. o, Basal blast resistance in NIL-Pigm and OsACS2-KO/NIL-Pigm, at 5 dpi with punch inoculation (TM21). p, Disease resistance of wild type and OsACS2-KO lines were shown, 5 dpi with TH12, NIPB served as a susceptible control. q, Met enhances rice blast resistance via ethylene biosynthesis. A Met solution (10 mM) or water was supplied from NIL-Pigm roots for 48 h and pre-treated with 10 μM AVG for 36 h before spray inoculation with rice blast (TM21). One-way ANOVA with Tukey’s test. Lesion areas (mean ± s.d.; n = 5, biologically independent samples), fungal growth (mean ± s.d.; n = 3, biologically independent samples). r, Protein levels of PICI1 in NIL-Pigm (left panel) and NIPB (right panel) plants after inoculation with M. oryzae at the indicated time points. Actin was detected as a loading control. The protein abundance was quantified using ImageJ and indicated under lanes. s, RNA expression of PICI1 was not significantly affected in late stage by blast infection (TH12 and TM21) in NIL-Pigm. For d, e, f, k, l, s, data are mean ± s.d. (n = 3, biologically independent samples). For h, i, j, o, p, data were analysed by two-tailed Student’s t-test. Lesion lengths or areas (mean ± s.d.; n = numbers of biologically independent samples in the graphs), fungal growth (mean ± s.d.; n = 3, biologically independent samples). Lowercase letters indicate statistical significance (P < 0.05) (a, m, q). Asterisks indicate significant differences (*P < 0.05, **P < 0.01) (b, c, g–j, o, p). Exact P values are provided in Supplementary Table 4 (a–c, g-j, m, o–q). The rice ACTIN1 served as an internal control (d–g, k, l, s). Scale bars, 1 cm (h–j, o–q). Experiments were repeated twice (d–g) three times with similar results (a–c, h–j, k–m, o-q, r, s).
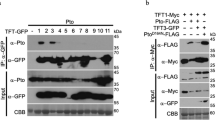
Extended Data Fig. 8 PigmR protects PICI1 from AvrPi9-mediated degradation.
a, His-PICI1 could be pulled down by seven effectors (AvrPWL2, AvrPi9, AvrPikC, AvrPia, AvrPizt, AvrPii and AvrPikD fused with MBP) in vitro. MBP alone served as a negative control. b, SLC verification of the interactions between PICI1 and the three blast fungal effectors, AvrPWL2, AvrPi9 and AvrPii in N. benthamiana. Note that AvrPikD/Pik-HMA and AvrPii/OsNADP-ME2 served as positive controls. c, Relative luciferase activity of protein-protein interactions was measured. Data are mean ± s.d. (n = 6, biologically independent samples). d, Western blot confirming protein expression in N. benthamiana. Note that 1-12 represented PICI1/AvrBAS4, PICI1/AvrPWL2, PICI1/AvrPi9, PICI1/AvrPita, PICI1/AvrSlp1, PICI1/AvrPikC, PICI1/AvrPia, PICI1/AvrPizt, PICI1/AvrPii, OsNADP-ME2/PICI1, PICI1/AvrPikD and Pik-HMA/AvrPikD, respectively. e–g, SLC assays showed that PICI1, but not PICI2, specifically interacted with AvrPWL2 (e), AvrPi9 (f) and AvrPii (g) in N. benthamiana. Fluorescence signal intensity is indicated. Protein expression in N. benthamiana was detected by western blot (e-g, down panel). h, Protein levels of endogenous PICI1 in AvrPi9-Flag (AvrPi9-OE), AvrPWL2-Flag (AvrPWL2-OE), AvrPizt-Flag (AvrPizt-OE), and AvrPii-Flag (AvrPii-OE), compared with NIL-Pigm. The protein abundance was quantified and indicated under lanes. i, Subcellular localization of AvrPi9 and co-localization of PICI1-YFP/AvrPizt-mCherry in rice protoplasts. j, Western blot analysis confirming expression of AvrPi9-mCherry or AvrPi9-YFP without free mCherry or YFP in rice protoplasts. k, BiFC confirms the PICI1/AvrPi9 interaction in rice protoplasts. PICI2 served as a negative control. l, Basal blast resistance of WT and AvrPi9-OE lines, 5 dpi with TM21. Scale bars, 1 cm. Data were analysed by two-tailed Student’s t-test. Lesion lengths (mean ± s.d.; n = numbers of biologically independent samples in the graphs), fungal growth (mean ± s.d.; n = 3, biologically independent samples). m, n, Induction of the PTI-related defence genes OsPAL1 (m) and KS4 (n) at 1 h after chitin incubation in WT and AvrPi9-OE. Two-tailed Student’s t-test (mean ± s.d.; n = 3, biologically independent samples). The rice ACTIN1 served as an internal control. o, Cell-free degradation shows AvrPi9FL-mediated PICI1-GFP degradation. Protein extracts were prepared from two-week-old PICI1-GFP/NIL-Pigm seedlings and then incubated with AvrPi9FL-mCherry or AvrPizt-mCherry expressed in N. benthamiana. p, Cell-free degradation shows the AvrPi9FL-mediated PICI1-GFP degradation in NIPB (left panel) and NIL-Pigm (right panel) with or without MG132. Protein extracts were prepared from PICI1-GFP/NIPB and PICI1-GFP/NIL-Pigm seedlings and then incubated with mCherry (upper panels) or AvrPi9-mCherry (lower panels) expressed in N. benthamiana. FL, full length (o, p), and we used the Avrs without signal peptide to make the constructs in the experiments unless otherwise indicated. q, Immunodetection of two independent transformants of TH12AvrPi9 using an anti-S-tag antibody, with its parent TH12 as a negative control. Full length of AvrPi9 tagged by S-tag was transformed into TH12 by Agrobacterium-mediated transformation. r, Pathogenicity test of the transformants with TH12 as a virulent control. s, Protein levels of PICI1 in NIPB (left panel) and NIL-Pigm (right panel) after inoculation with TH12 (upper panel) or TH12AvrPi9 (lower panel). The protein abundance was quantified using ImageJ and indicated under lanes. Ponceau S staining (o–q) or Actin (d–h, j, s) served as loading control. Scale bars, 5 µm (i, k). Asterisks represent significant difference (*P < 0.05, **P < 0.01) (l-n). Exact P values are provided in Supplementary Table 4 (l–n). Experiments were repeated twice (a, d, h, j, q, s) or three times (b, c, e–g, i, k-p, r) with similar results.
Extended Data Fig. 9 PigmR and Pizt protect PICI1 from Avrs binding in a competitive manner.
a, PICI1-interacting effectors including AvrPi9, AvrPWL2 and AvrPii can not activate PigmR-mediated cell death. Relative luciferase activity was measured after transformation with different effectors in NIPB or NIL-Pigm. Empty vector (EV) and MoCDIP1 were used as negative and positive control, respectively. One-way ANOVA with Tukey’s test (mean ± s.d.; n = 3, biologically independent experiments). Different letters indicate significant difference at P < 0.05. b, Western blot analysis confirming expression of proteins in rice protoplasts. Note that 1-5 and 6-10 represented MoCDIP1, EV, AvrPi9, AvrPWL2 and AvrPii in NIPB and NIL-Pigm, respectively. The asterisks indicate the target proteins. c, Increased protein accumulation of PigmR in PigmR-7Myc-His/NIPB after inoculation with TH12 or TH12AvrPi9 with water as a control at the indicated time points. d, PICI1-PigmR interaction is enhanced by M. oryzae infection. Proteins were prepared from transgenic plants PICI1-GFP/NIL-Pigm and PigmR-7Myc-His/NIPB after inoculation with TH12 or water as a control (36 hpi). e, A SLC assay shows that the PigmR disrupts PICI1-AvrPi9 interaction in N. benthamiana. Relative luciferase activity of protein-protein interactions was measured (lower panel). Two-tailed Student’s t-test (mean ± s.d.; n = 6, biologically independent samples). The asterisk represents significant difference (**P < 0.01). f, Western blot confirming protein expression in N. benthamiana. Note that left and right represent PICI1/AvrPi9/GFP and PICI1/AvrPi9/PigmR combination, respectively. g, PigmR-CC outcompetes AvrPi9 for PICI1 binding in vitro. h, PigmR-CC outcompetes AvrPi9 for PICI1 binding in rice protoplasts. i, j, SLC assays show that the PigmR-CC also disrupts PICI1/AvrPWL2 (i) and PICI1/AvrPii (j) interactions in N. benthamiana. Protein expression in N. benthamiana was detected by western blot (lower panel) (i, j). k, l, PigmR-CC outcompetes AvrPii (k) or AvrPWL2 (l) for PICI1 binding in rice protoplasts. The asterisks indicate the target proteins (l). m, n, Protein accumulation of PICI1 and OsMETS in Ky-Pi9 (m) and ZH11 (n) plants after inoculation with avirulent strain Guy11 (m) or YN2 (n) at the indicated time points. o, Protein accumulation of PICI1 in ZH11 and Pizt-KO/ZH11 plants after inoculation with blast strain CH131 (avirulent to Pizt) at the indicated time points. p, SLC assays show that the Pizt-CC also disrupts PICI1/AvrPi9 (left panel), PICI1/AvrPWL2 (middle panel) and PICI1/AvrPii (right panel) interactions in N. benthamiana. Protein expression in N. benthamiana was detected by western blot (lower panel). Actin (b, c, f, i, j, m–o) or Ponceau S staining (p) used as loading control. The protein abundance was quantified using ImageJ and indicated under lanes (c, m-o). Fluorescence signal intensity is indicated (e, i, j, p). Exact P values are provided in Supplementary Table 4 (a, e). Experiments were repeated twice (b, d, f, g, h, k-p) or three times (a, c, e, i, j) with similar results.
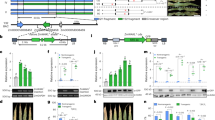
Extended Data Fig. 10 PICI1 jap allele shows higher PICI1 expression and increased basal defence, and acetylation of PICI1 promotes its degradation.
a, Twenty-seven SNPs in the PICI1 promoter distribute in different rice varieties based on the 3,000 Rice Genome database33. SNP = 0 indicates the same promoter sequence of PICI1 with NIPB, which was used as the referee. SNP > 0 indicated at least one base change. b, Distribution of the 27 SNPs in japonica and indica rice based on the 3,000 Rice Genome Project database. c, FST of PICI1 and flanking regions between different rice groups. d, Induction of ProPICI1ind-LUC and ProPICI1jap-LUC in rice protoplasts by chitin. LUC activity was measured by normalizing to REN signal. e, japonica rice varieties showed a general higher induction of PICI1 than indica varieties by M. oryzae cocktail at 3 hpi. In box plots, the centre line represents the median, box edges delimit lower and upper quartiles and whiskers show the highest and lowest data points. Two-tailed Student’s t-test (mean ± s.d.; n = numbers of biologically independent samples in the graphs). f, Pathotype test of different M. oryzae strains in two-week-old seedlings of Huajingxian74 using spraying inoculation, indicating YN2 and 99-30-1 virulent toward Huajingxian74. g, Induction of PICI1 at indicated times by M. oryzae infection in Huajingxian74 and CSSL. h, Induction of the defence gene KS4 after blast inoculation in Huajingxian74 and CSSL plants. i, Pathotype test of M. oryzae isolate and Guy11 (avirulent) and TM21 (virulent) toward Pi9-containing rice plant (Ky-Pi9) with punch inoculation. j, Increased acetylation of PICI1 after blast infection. The protein extracts from PICI1-GFP/NIL-Pigm plants challenged with M. oryzae (TM21) or water for 36 h were immunoprecipitated with Ac-K (Acetylated-lysine) or GFP antibody and analysed using anti-GFP or anti-Ac-K antibody, respectively. k, A SLC assay shows the reduced interaction of PICI1K160Q/PigmR-CC and PICI1K160Q/OsMETS1 in comparison with PICI1 in N. benthamiana. The protein and transcript levels of cLuc-PICI1 and cLuc-PICI1K160Q were determined by immunoblot and semi-quantitative PCR. Actin was detected as a loading control (j, k). The rice ACTIN1 served as an internal control (e, g, h). Two-tailed Student’s t-test (mean ± s.d.; n = 3, biologically independent samples) (d, g, h). Asterisks indicate significant differences (*P < 0.05, **P < 0.01) (d, e, g, h). Exact P values are provided in Supplementary Table 4 (d, e, g, h). Scale bars, 1 cm (f, i). Experiments were repeated twice (e, g, h) or three times (d, f, i-k) with similar results.
Supplementary information
Supplementary Figure 1
Uncropped blots and gel images.
Supplementary Table 1
MS/MS and statistical analysis.
Supplementary Table 2
Natural variation analysis for PICI1.
Supplementary Table 3
Primers used in this study.
Supplementary Table 4
Statistical summary—a summary of all statistical analysis is provided. Source data for all of the statistical analysis can be found in the Source Data files.
Source data
Rights and permissions
About this article
Cite this article
Zhai, K., Liang, D., Li, H. et al. NLRs guard metabolism to coordinate pattern- and effector-triggered immunity. Nature 601, 245–251 (2022). https://doi.org/10.1038/s41586-021-04219-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04219-2
This article is cited by
-
Early molecular events in the interaction between Magnaporthe oryzae and rice
Phytopathology Research (2024)
-
Deciphering Aphanomyces euteiches-pea-biocontrol bacterium interactions through untargeted metabolomics
Scientific Reports (2024)
-
The OsSGS3-tasiRNA-OsARF3 module orchestrates abiotic-biotic stress response trade-off in rice
Nature Communications (2023)
-
Inactivation of the antidiabetic drug acarbose by human intestinal microbial-mediated degradation
Nature Metabolism (2023)
-
New insights from a plant immune receptor may help design of disease resistant crops
Science China Life Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.