Abstract
The σ2 receptor has attracted intense interest in cancer imaging1, psychiatric disease2, neuropathic pain3,4,5 and other areas of biology6,7. Here we determined the crystal structure of this receptor in complex with the clinical candidate roluperidone2 and the tool compound PB288. These structures templated a large-scale docking screen of 490 million virtual molecules, of which 484 compounds were synthesized and tested. We identified 127 new chemotypes with affinities superior to 1 μM, 31 of which had affinities superior to 50 nM. The hit rate fell smoothly and monotonically with docking score. We optimized three hits for potency and selectivity, and achieved affinities that ranged from 3 to 48 nM, with up to 250-fold selectivity versus the σ1 receptor. Crystal structures of two ligands bound to the σ2 receptor confirmed the docked poses. To investigate the contribution of the σ2 receptor in pain, two potent σ2-selective ligands and one potent σ1/σ2 non-selective ligand were tested for efficacy in a mouse model of neuropathic pain. All three ligands showed time-dependent decreases in mechanical hypersensitivity in the spared nerve injury model9, suggesting that the σ2 receptor has a role in nociception. This study illustrates the opportunities for rapid discovery of in vivo probes through structure-based screens of ultra large libraries, enabling study of underexplored areas of biology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The coordinates and structure factors for PB28-bound σ2, roluperidone-bound σ2, Z1241145220-bound σ2, Z4857158944-bound σ2 and cholesterol-bound σ2 have been deposited in the PDB with accession codes 7M93, 7M94, 7M95, 7M96 and 7MFI, respectively. The identities of the compounds docked in this study are freely available from the ZINC database (http://zinc15.docking.org) and active compounds may be purchased from Enamine. Any other data relating to this study are available from the corresponding authors on reasonable request. Source data are provided with this paper.
Code availability
DOCK 3.7 is freely available for non-commercial research http://dock.compbio.ucsf.edu/DOCK3.7/. A web-based version is available at http://blaster.docking.org/.
References
Waarde, A. V. et al. Potential applications for sigma receptor ligands in cancer diagnosis and therapy. Biochim. Biophys. Acta 1848, 2703–2714 (2015).
Harvey, P. D. et al. Effects of roluperidone (MIN-101) on two dimensions of the negative symptoms factor score: reduced emotional experience and reduced emotional expression. Schizophr. Res. 215, 352–356 (2020).
Sahn, J. J., Mejia, G. L., Ray, P. R., Martin, S. F. & Price, T. J. Sigma 2 receptor/Tmem97 agonists produce long lasting antineuropathic pain effects in mice. ACS Chem. Neurosci. 8, 1801–1811 (2017).
Intagliata, S. et al. Discovery of a highly selective sigma-2 receptor ligand, 1-(4-(6,7-dimethoxy-3,4-dihydroisoquinolin-2(1H)-yl)butyl)-3-methyl-1H-benzo[d]imidazol-2(3H)-one (CM398), with drug-like properties and antinociceptive effects in vivo. AAPS J. 22, 94 (2020).
Quadir, S. G. et al. The Sigma-2 receptor / transmembrane protein 97 (σ2R/TMEM97) modulator JVW-1034 reduces heavy alcohol drinking and associated pain states in male mice. Neuropharmacology 184, 108409 (2021).
Grundman, M. et al. A phase 1 clinical trial of the sigma‐2 receptor complex allosteric antagonist CT1812, a novel therapeutic candidate for Alzheimer’s disease. Alzheimers Dement. 5, 20–26 (2018).
Riad, A. et al. Sigma-2 receptor/TMEM97 and PGRMC-1 increase the rate of internalization of LDL by LDL receptor through the formation of a ternary complex. Sci. Rep. 8, 16845 (2018).
Abate, C. et al. PB28, the sigma-1 and sigma-2 receptors modulator with potent anti–SARS-CoV-2 activity: a review about its pharmacological properties and structure affinity relationships. Front. Pharmacol. 11, 589810 (2020).
Shields, S. D., Eckert, W. A. & Basbaum, A. I. Spared nerve injury model of neuropathic pain in the mouse: a behavioral and anatomic analysis. J. Pain 4, 465–470 (2003).
Hellewell, S. B. et al. Rat liver and kidney contain high densities of σ1 and σ2 receptors: characterization by ligand binding and photoaffinity labeling. Eur. J. Pharmacol. 268, 9–18 (1994).
Hellewell, S. B. & Bowen, W. D. A sigma-like binding site in rat pheochromocytoma (PC12) cells: decreased affinity for (+)-benzomorphans and lower molecular weight suggest a different sigma receptor form from that of guinea pig brain. Brain Res. 527, 244–253 (1990).
Hanner, M. et al. Purification, molecular cloning, and expression of the mammalian sigma1-binding site. Proc. Natl. Acad. Sci. U.S.A. 93, 8072–8077 (1996).
Langa, F. et al. Generation and phenotypic analysis of sigma receptor type I (sigma1) knockout mice. Eur. J. Neurosci. 18, 2188–2196 (2003).
Alon, A. et al. Identification of the gene that codes for the σ2 receptor. Proc. Natl. Acad. Sci. U.S.A. 114, 7160–7165 (2017).
Ebrahimi-Fakhari, D. et al. Reduction of TMEM97 increases NPC1 protein levels and restores cholesterol trafficking in Niemann-pick type C1 disease cells. Hum. Mol. Genet. 25, 3588–3599 (2016).
Bartz, F. et al. Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 10,75 (2009).
Sanchez-Pulido, L. & Ponting, C. P. TM6SF2 and MAC30, new enzyme homologs in sterol metabolism and common metabolic disease. Front. Genet. 5, 439 (2014).
Mahdessian, H. et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc. Natl. Acad. Sci. U.S.A. 111, 8918 (2014).
Vilner, B. J., John, C. S. & Bowen, W. D. Sigma-1 and sigma-2 receptors are expressed in a wide variety of human and rodent tumor cell lines. Cancer Res. 55, 408–413 (1995).
Scott, L. L. et al. Small molecule modulators of σ2R/Tmem97 reduce alcohol withdrawal-induced behaviors. Neuropsychopharmacology 43, 1867–1875 (2018).
Vázquez-Rosa, E. et al. Neuroprotective efficacy of a sigma 2 receptor/TMEM97 modulator (DKR-1677) after traumatic brain injury. ACS Chem. Neurosci. 10, 1595–1602 (2019).
Stein, R. M. et al. Virtual discovery of melatonin receptor ligands to modulate circadian rhythms. Nature 579, 609–614 (2020).
Schuller, M. et al. Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking. Sci. Adv. 7, eabf8711 (2021).
Schmidt, H. R. & Kruse, A. C. The molecular function of σ receptors: past, present, and future. Trends Pharmacol. Sci. 40, 636–654 (2019).
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009).
Long, T. et al. Structural basis for human sterol isomerase in cholesterol biosynthesis and multidrug recognition. Nat. Commun. 10, 2452 (2019).
Audet, M. & Stevens, R. C. Emerging structural biology of lipid G protein‐coupled receptors. Protein Sci. 28, 292–304 (2019).
Schmidt, H. R. et al. Crystal structure of the human σ1 receptor. Nature 532, 527–530 (2016).
Hubler, Z. et al. Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination. Nature 560, 372–376 (2018).
Lyu, J. et al. Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019).
Fischer, A., Smieško, M., Sellner, M. & Lill, M. A. Decision making in structure-based drug discovery: visual inspection of docking results. J. Med. Chem. 64, 2489–2500 (2021).
Cendán, C. M., Pujalte, J. M., Portillo-Salido, E., Montoliu, L. & Baeyens, J. M. Formalin-induced pain is reduced in σ1 receptor knockout mice. Eur. J. Pharmacol. 511, 73–74 (2005).
Puente, B. D. L. et al. Sigma-1 receptors regulate activity-induced spinal sensitization and neuropathic pain after peripheral nerve injury. Pain 145, 294–303 (2009).
Cendán, C. M., Pujalte, J. M., Portillo-Salido, E. & Baeyens, J. M. Antinociceptive effects of haloperidol and its metabolites in the formalin test in mice. Psychopharmacology 182, 485–493 (2005).
Romero, L. et al. Pharmacological properties of S1RA, a new sigma‐1 receptor antagonist that inhibits neuropathic pain and activity‐induced spinal sensitization. Br. J. Pharmacol. 166, 2289–2306 (2012).
Bruna, J. et al. Efficacy of a novel sigma-1 receptor antagonist for oxaliplatin-induced neuropathy: a randomized, double-blind, placebo-controlled phase IIa clinical trial. Neurotherapeutics 15, 178–189 (2018).
Vela, J. M., Merlos, M. & Almansa, C. Investigational sigma-1 receptor antagonists for the treatment of pain. Expert Opin. Investig. Drugs 24, 883–896 (2015).
Kooistra, A. J. et al. GPCRdb in 2021: integrating GPCR sequence, structure and function. Nucleic Acids Res. 49, D335–D343 (2021).
Hauser, A. S. et al. Pharmacogenomics of GPCR drug targets. Cell 172, 41–54 (2018).
Kroeze, W. K. et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 22, 362–369 (2015).
Nastasi, G. et al. S2RSLDB: a comprehensive manually curated, internet-accessible database of the sigma-2 receptor selective ligands. J. Cheminformatics 9, 3 (2017).
Huang, X.-P. et al. Allosteric ligands for the pharmacologically dark receptors GPR68 and GPR65. Nature 527, 477–483 (2015).
Wang, S. et al. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature 555, 269–273 (2018).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012).
Gordon, D. E. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Kabsch, W. XDS. Acta Crystallogr. D 66,132 (2010).
Alford, R. F. et al. An integrated framework advancing membrane protein modeling and design. PLoS Comput. Biol. 11, e1004398 (2015).
Mccoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40,674 (2007).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D 75, 861–877 (2019).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
The PyMOL Molecular Graphics System v.2.5 (Schrödinger).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Chu, U. B. & Ruoho, A. E. Sigma receptor binding assays. Curr. Protoc. Pharmacol. 71, 1.34.1–1.34.21 (2015).
Weiner, S. J. et al. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 106, 765–784 (1984).
Meng, E. C., Shoichet, B. K. & Kuntz, I. D. Automated docking with grid‐based energy evaluation. J. Comput. Chem. 13, 505–524 (1992).
Gallagher, K. & Sharp, K. Electrostatic contributions to heat capacity changes of DNA–ligand binding. Biophys. J. 75, 769–776 (1998).
Mysinger, M. M. & Shoichet, B. K. Rapid context-dependent ligand desolvation in molecular docking. J. Chem. Inf. Model. 50, 1561–1573 (2010).
Stein, R. M. et al. Property-unmatched decoys in docking benchmarks. J. Chem. Inf. Model. 61, 699–714 (2021).
Gu, S., Smith, M. S., Yang, Y., Irwin, J. J. & Shoichet, B. K. Ligand strain energy in large library docking. J. Chem. Inf. Model. 61, 4331–4341 (2021).
Carpenter, B. et al. Stan: a probabilistic programming language. J. Stat. Softw. 76, 1–32 (2017).
Bürkner, P.-C. brms: an R package for Bayesian multilevel models using Stan. J. Stat. Softw. 80, 1–28 (2017).
Gelman, A., Goodrich, B., Gabry, J. & Vehtari, A. R-squared for Bayesian regression models. Am. Statistician 73, 307–309 (2018).
Kay, M. tidybayes: tidy data and geoms for Bayesian models. https://doi.org/10.5281/zenodo.1308151 (Zenodo, 2020).
Wickham, H. ggplot2: elegant graphics for data analysis (Springer, 2016).
Wickham, H. et al. Welcome to the tidyverse. J. Open Source Softw. 4, 1686 (2019).
R Core Team. R: a Language and Environment for Statistical Computing. http://www.R-project.org (R Foundation for Statistical Computing, 2018).
Skubic, C., Vovk, I., Rozman, D. & Križman, M. Simplified LC-MS method for analysis of sterols in biological samples. Molecules 25, 4116 (2020).
Longo, P. A., Kavran, J. M., Kim, M.-S. & Leahy, D. J. Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol. 529, 227–240 (2013).
Besnard, J. et al. Automated design of ligands to polypharmacological profiles. Nature 492, 215–220 (2012).
Scherrer, G. et al. Dissociation of the opioid receptor mechanisms that control mechanical and heat pain. Cell 137, 1148–1159 (2009).
Muralidharan, A. et al. Identification and characterization of novel candidate compounds targeting 6‐ and 7‐transmembrane μ‐opioid receptor isoforms. Br. J. Pharmacol. 178, 2709–2726 (2021).
Chaplan, S. R., Bach, F. W., Pogrel, J. W., Chung, J. M. & Yaksh, T. L. Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods 53, 55–63 (1994).
Solorzano, C. et al. Primary afferent and spinal cord expression of gastrin-releasing peptide: message, protein, and antibody concerns. J. Neurosci. 35, 648–657 (2015).
Notredame, C., Higgins, D. G. & Heringa, J. T-coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 (2000).
Liebschner, D. et al. Polder maps: improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D 73, 148–157 (2017).
Acknowledgements
Funding to support this research was provided by NIH grant R01GM119185, the Vallee Foundation and the Sanofi iAwards program (A.C.K.); by DARPA grant HR0011-19-2-0020 and NIH grant R35GM122481 (B.K.S.); and by grant GM133836 (J.J.I.). GM/CA @ APS has been funded by the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006 and P30GM138396). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357. The Eiger 16M detector at GM/CA-XSD was funded by NIH grant S10 OD012289. We thank K. Arnett and the Harvard Center for Macromolecular Interactions for support with biophysical experiments including circular dichroism and SEC–MALS, and C. Vidoudez and the Harvard Center for Mass Spectrometry for performing mass spectrometry analysis of sterols. Molecular graphics and analyses were performed with UCSF Chimera, which was developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
Author information
Authors and Affiliations
Contributions
A.A. performed cloning, mutagenesis, protein purification, SEC–MALS experiments, circular dichroism measurements, crystallization, X-ray data collection and processing, structure determination and refinement, radioligand binding, yeast complementation experiments and the sterol isomerization enzymatic assay. J.L. conducted the docking, chemoinformatics analyses, docking energy analysis and ligand picking, assisted in the latter by T.A.T. and B.K.S. J.M.B. conducted and analysed the mouse allodynia experiments with assistance from V.C., as well as the receptor expression experiments, which were supervised and co-analysed by A.I.B. M.J.O. conducted the Bayesian analysis of docking scores versus hit rates. C.M.W. tested molecules for activity against the μOR. X.-P.H. and Y.L. tested compounds against the GPCRome and other off-targets, with supervision from B.L.R. Y.S.M. supervised the synthesis of molecules from the virtual library. D.S.R. participated in the creation of Enamine REAL library. J.J.I. was responsible for the building of the version of the ZINC library that was docked. A.C.K., B.K.S. and A.I.B. supervised the project. The manuscript was written by A.A., J.L., B.K.S. and A.C.K. with input from other authors.
Corresponding authors
Ethics declarations
Competing interests
A.C.K. is a founder and consultant for biotechnology companies Tectonic Therapeutic and Seismic Therapeutic, as well as the Institute for Protein Innovation, a non-profit research institute. B.K.S. is a founder of Epiodyne, a company active in analgesia, and of Blue Dolphin, which undertakes fee-for-service ligand-discovery.
Additional information
Peer review information Nature thanks Kevin Jones and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Characterization of the σ2 receptor.
a, SEC–MALS of the human σ2 receptor. The σ2 receptor was run either without ligand or with 1 µM of the indicated ligand. Lines indicate calculated total mass (grey), detergent micelle (purple), and protein (blue). b, Sequence alignment between the human and bovine σ2 protein sequences performed using T-coffee76. Residues that line the binding pocket are marked in red. c, Circular dichroism analysis of the bovine σ2 receptor alone (black) or with the indicated ligand. Data is representative of multiple experiments. d, Circular dichroism melting curves of the bovine σ2 receptor. Temperature was raised from 20 °C to 90 °C and molar ellipticity was measured at 222 nm. Protein was incubated either with or without indicated ligand at 12 µM. Melting temperatures for each measurement are indicated with a circle. Data is representative of multiple experiments e, SEC of the bovine σ2 receptor. Blue trace is after proteolytic tag removal. Red trace is protein applied on size exclusion after reapplying the tag-free protein on affinity resin to remove proteins with intact tags. The trace presented is representative of multiple purifications. f, Analysis of receptor purity after the second SEC using SDS–PAGE. Grey rectangle in e represents fractions chosen for analysis. The SDS-PAGE presented here is representative of multiple purifications. See Source Data for uncropped version. g, Crystals of bovine σ2 receptor in the lipidic cubic phase. h, Aspartate 56 (D56) is important for receptor structure but not for ligand binding. A tight network of hydrogen bonds that bridges extracellular loop 1 to TM4 is depicted with black dashed lines. i, Electron density maps for the various ligands. Polder maps77 were calculated in PHENIX. Maps are contoured at a level of 3 σ. j, View of cholesterol-binding pose, showing contacts with other binding pocket residues. Hydrogen bonds are marked with black dashed lines. k, Yeast complementation assay. A ΔERG2 yeast strain was transformed with plasmids harbouring the indicated genes. Yeast cells were grown to logarithmic phase and diluted to OD600 of 0.1, and then further diluted in a five-fold serial dilution series. Two microliters of each dilution were spotted on plates. Yeast cells were grown either in permissive conditions of no cycloheximide or in the restrictive conditions of 50 ng/ml cycloheximide, which requires functional Δ8-9 sterol isomerase activity for viability. ERG2 and EBP can act as sterol isomerases and rescue the growth of ΔERG2 S. cerevisiae whereas the σ2 receptor, the σ1 receptor, or any other member of the EXPERA family cannot. l, EBP can catalyse the conversion of zymostenol to lathosterol whereas σ2 cannot. Standards are in dark grey. EBP converts zymostenol to lathosterol (apricot) but does not convert lathosterol to zymostenol (dark red). The σ2 receptor does not convert lathosterol to zymostenol (dark blue) or zymostenol to lathosterol (light purple). Structures of zymostenol and lathosterol are depicted below the traces
Extended Data Fig. 2 Comparisons of the distribution of docking scores.
a–d, The distribution of docking scores of tested molecules for hit rate curves against σ2 (left column) and D4 (right column) receptors. All tested molecules are grouped based on docking score bins. The distributions are shown in box plots for a, net electrostatic energy, b, ligand desolvation energy, c, van der Waals (vdW) energy and d, delta ligand desolvation energy after recalculating atomic desolvation energy based on the docked pose. e–h, Comparison of hit rates and affinities achieved by combined docking score and human inspection and these achieved by docking score alone. e, Overall hit rates for selecting compounds from the first 3 scoring bins by each strategy: human prioritization and docking score (orange), or docking score alone (blue). Hit rate is the ratio of active compounds/tested compounds; the raw numbers appear at the top of each bar. f, Hit rates for selecting compounds at different scoring ranges by each strategy: human prioritization and docking score (orange) or docking score alone (blue). g, Distribution of the binding affinity level among the hits from e (top panel). We measured competition binding curves for 14 docking hits from human prioritization and docking score, and 7 hits from the docking score alone. These are divided into three affinity ranges: <5 nM; 5 nM–50 nM; >50 nM; Distribution of the binding affinity level among the hits from all different scoring ranges (bottom panel). We measured competition binding curves for 14 docking hits from human prioritization and docking score, and 17 hits from the docking score alone. h. Hit-rate curve comparison with/without human picks. The hit rate without human picks at the top plateau is 39% and at the bottom plateau is 0%, and the docking score (dock50) and slope at the maximum (slope50) are -46.5 kcal mol−1 and -3.5% per kcal mol−1, respectively
Extended Data Fig. 3 Analogues of σ2 receptor ligands and the effect of a structural water molecule.
a–c, Initial hits and selected analogues of σ2 receptor ligands. Competition binding curves on the top panel, 2D drawings of compounds are on the bottom panel. Parent compound is indicated by grey background. Points shown as mean ± s.e.m. from three technical replicates. a, Parent compound ZINC548355486 and its three potent analogues. b, Parent compound ZINC895657866 and its three potent analogues. c, Parent compound ZINC450573233 and its three potent analogues. d–f, The binding site of the σ2 receptor contains a structural water. d, Water coordination at the binding site of the σ2 receptor. Water molecule is depicted as a red sphere. Hydrogen bonds are indicated by black dashed lines. e, Saturation binding curve to measure the dissociation constant (Kd) of [3H]DTG for the various mutants of σ2 receptor meant to disrupt water coordination. Residues proximal to the structural water were chosen for mutation. Residues were mutated to the indicated amino acid. Points shown as mean ± s.e.m. from three technical replicates. f, Competition binding measurement of affinity of Z1241145220 in various mutants of σ2. Points shown as mean ± s.e.m. from three technical replicates
Extended Data Fig. 4 Effect of systemic σ receptor ligands on motor behaviour.
a, Response of mice to a von Frey filament after spared nerve injury (SNI). All five ligands are compared to their respective vehicles (PD-144418 10 mg/kg (n = 5) and 30 mg/kg (n = 5) vs. kolliphor (n = 5), one-way ANOVA, F(2, 12) = 7.49, p = 0.008; Z4446724338 10 mg/kg (n = 10) and 20 mg/kg (n = 5) vs cyclodextrin (n = 10), one-way ANOVA, F(2, 22) = 25.12, p < 0.001; Z4857158944 10 mg/kg (n = 5) and 20 mg/kg (n = 5) vs cyclodextrin (n = 10), one-way ANOVA, F(2, 17) = 5.10, p = 0.02; Z1665845742 10 mg/kg (n = 10) and 20 mg/kg (n = 5) and PB28 30 mg/kg (n = 10) vs saline (n = 10), one-way ANOVA, F(3, 31) = 6.18, p = 0.002; asterisks define individual group differences to respective vehicle control using Dunnett’s multiple comparisons Post-hoc test; ns = not significant, * p < 0.05, ** p < 0.01, *** p < 0.001). Data shown are mean ± s.e.m. Data for higher doses and vehicles is replotted from Fig. 4. b, No sedation or motor impairment on the rotarod was observed after drug treatments compared to vehicle at 1 h (Z1665845742 10 mg/kg (n = 5) and Z4857158944 20 mg/kg (n = 5) vs saline (n = 5), one-way ANOVA, F(2, 12) = 1.04, p = 0.38; Z4446724338 10 mg/kg (n = 5) vs kolliphor (n = 5), unpaired two-tailed Student’s t-test, t(8) = 0.47, p = 0.65) or 24 h post-injection (Z1665845742 10 mg/kg (n = 5) and Z4857158944 20 mg/kg (n = 5) vs saline (n = 5), one-way ANOVA, F(2, 12) = 0.45, p = 0.65; Z4446724338 10 mg/kg (n = 5) vs kolliphor (n = 5), unpaired two-tailed Student’s t-test, t(8) = 0.72, p = 0.49); ns = not significant. Data shown are means ± s.e.m. c, Response of SNI mice to a von Frey filament after repeated injections of Z4446724338 10 mg/kg (n = 5). Mechanical thresholds were assessed 1 h and 24 h after four separate injections. Data shown are means ± s.e.m. normalized to each mouse’s SNI baseline. d, Response of SNI mice to a von Frey filament after repeated injections of Z4857158944 10 mg/kg (n = 5). Mechanical thresholds were assessed 1 h and 24 h after four separate injections. Data shown are means ± s.e.m. normalized to each mouse’s SNI baseline. e. Quantification of the expression levels of Sigmar1 (σ1) and Tmem97 (σ2) in wildtype (WT) and SNI mice detected by in situ hybridization (n = 3 mice per group). Representative images can be found in panel f. Data shown are mean ± s.e.m.; unpaired two-tailed Student’s t-test— Sigmar1: t(4) = 0.5, p = 0.64; Tmem97: t(4) = 1.0, p = 0.37; ns = not significant. AU = arbitrary units. f, in situ hybridization of mouse dorsal root ganglion (DRG) sections for Sigmar1 (σ1) and Tmem97 (σ2) genes illustrates expression in myelinated (Nefh-positive; blue) and unmyelinated (Acpp-positive; red) subsets of sensory neurons and no change after SNI
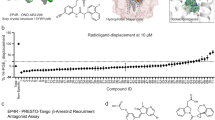
Extended Data Fig. 5 Off-target profiling of Z4446724338, Z1665845742 and Z4857158944.
a–c, TANGO screens against a panel of 320 GPCRs for the indicated σ2 ligand. a, Z4446724338, b, Z1665845742, c, Z4857158944. d, GloSensor μOR-mediated cAMP inhibition (Gi activation) by DAMGO, Z4446724338, Z1665845742, and Z4857158944. e, f, Follow-up does-response curves for pain-related receptors that showed activation in a–c. e, Z4446724338 and Z1665845742 against 5HT1A. f, Z4857158944 against κOR. Data shown are means ± s.e.m
Extended Data Fig. 6 Paw withdrawal thresholds.
a, Paw withdrawal thresholds (PWT) before (blue bar) and after (red bar) SNI, as well as after SNI + treatment (purple bar). For easier visualization of individual data points, data was also plotted without the pre-SNI baseline. Data are the same as in Fig. 4b and Extended Data Fig. 4a, but without the normalization to the individual post-SNI baselines and are expressed as mean ± s.e.m.; mice per group: saline (n = 10); cyclodextrin (n = 10); kolliphor (n = 5); PB28 30 mg/kg (n = 10); PD-144418 10 mg/kg (n = 5) and 30 mg/kg (n = 5); Z4446724338 10 mg/kg (n = 10) and 20 mg/kg (n = 5); Z1665845742 10 mg/kg (n = 5) and 20 mg/kg (n = 5); Z4857158944 10 mg/kg (n = 5) and 20 mg/kg (n = 5); unpaired two-tailed Student’s t-test. b, PWTs 1 h, 24 h, and 48 h after saline or drug treatment. Data are the same as in Fig. 4c, but without the normalization to the individual post-SNI baselines, and are expressed as mean ± s.e.m. Significance levels determined using Dunnett’s multiple comparisons Post-hoc test reflect the difference between Z4446724338 and saline for simplicity (two-way ANOVA; time x treatment interaction: F(8, 80) = 2.4, p = 0.02; time: F(2, 74) = 5.2, p = 0.009; treatment: F(4, 40) = 3.3, p = 0.02; four treatment groups (n = 10) except PD-144418 (n = 5); ns = not significant. c, Response of SNI mice to a von Frey filament after repeated injections of Z4446724338 10 mg/kg (n = 5). Mechanical thresholds were assessed 1 h and 24 h after four separate injections. Data shown are paw withdrawal thresholds in grams, expressed as mean ± s.e.m. d, Response of SNI mice to a von Frey filament after repeated injections of Z4857158944 10 mg/kg (n = 5). Mechanical thresholds were assessed 1 h and 24 h after four separate injections. Data shown are paw withdrawal thresholds in grams, expressed as mean ± s.e.m
Supplementary information
Supplementary Information
This file contains Supplementary Tables 2–5, Supplementary Figs. 1–8 and details regarding the Synthetic Procedures and Chemical Characterization Data.
Supplementary Table 1
Supplementary Table listing all the compounds tested in the manuscript.
Rights and permissions
About this article
Cite this article
Alon, A., Lyu, J., Braz, J.M. et al. Structures of the σ2 receptor enable docking for bioactive ligand discovery. Nature 600, 759–764 (2021). https://doi.org/10.1038/s41586-021-04175-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04175-x
This article is cited by
-
Exploring the combinatorial explosion of amine–acid reaction space via graph editing
Communications Chemistry (2024)
-
Increased rigidity and bioisosteric replacement in the design, synthesis and preliminary evaluation of novel, functionalized 3,3-dialkyl-γ-butyrolactones as sigma-2 ligands
Medicinal Chemistry Research (2024)
-
Modeling the expansion of virtual screening libraries
Nature Chemical Biology (2023)
-
Computational approaches streamlining drug discovery
Nature (2023)
-
Structure-based discovery of novel P-glycoprotein inhibitors targeting the nucleotide binding domains
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.