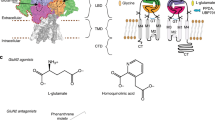
Abstract
Ketamine is a non-competitive channel blocker of N-methyl-d-aspartate (NMDA) receptors1. A single sub-anaesthetic dose of ketamine produces rapid (within hours) and long-lasting antidepressant effects in patients who are resistant to other antidepressants2,3. Ketamine is a racemic mixture containing equal parts of (R)- and (S)-ketamine, with the (S)-enantiomer having greater affinity for the NMDA receptor4. Here we describe the cryo-electron microscope structures of human GluN1–GluN2A and GluN1–GluN2B NMDA receptors in complex with S-ketamine, glycine and glutamate. Both electron density maps uncovered the binding pocket for S-ketamine in the central vestibule between the channel gate and selectivity filter. Molecular dynamics simulation showed that S-ketamine moves between two distinct locations within the binding pocket. Two amino acids—leucine 642 on GluN2A (homologous to leucine 643 on GluN2B) and asparagine 616 on GluN1—were identified as key residues that form hydrophobic and hydrogen-bond interactions with ketamine, and mutations at these residues reduced the potency of ketamine in blocking NMDA receptor channel activity. These findings show structurally how ketamine binds to and acts on human NMDA receptors, and pave the way for the future development of ketamine-based antidepressants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The cryo-EM maps and structure coordinates for GluN1–GluN2A and GluN1–GluN2B receptors have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-31308 and EMD-31309, and in the Protein Data Bank under accession numbers 7EU7 and 7EU8, respectively. The structures of S-ketamine and R-ketamine are accessible from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) under compound CIDs 182137 and 644025, respectively. The human missense mutation c.1925T>G (L642R) in GRIN2A was retrieved from the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) under variation ID 985631. Additional data that support the findings of this study are available from the corresponding author upon request.
Change history
14 October 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41586-021-04038-5
References
Autry, A. E. et al. NMDA receptor blockade at rest triggers rapid behavioural antidepressant responses. Nature 475, 91–95 (2011).
Berman, R. M. et al. Antidepressant effects of ketamine in depressed patients. Biol. Psychiatry 47, 351–354 (2000).
Fava, M. et al. Double-blind, placebo-controlled, dose-ranging trial of intravenous ketamine as adjunctive therapy in treatment-resistant depression (TRD). Mol. Psychiatry 25, 1592–1603 (2020).
Ebert, B., Mikkelsen, S., Thorkildsen, C. & Borgbjerg, F. M. Norketamine, the main metabolite of ketamine, is a non-competitive NMDA receptor antagonist in the rat cortex and spinal cord. Eur. J. Pharmacol. 333, 99–104 (1997).
Malhi, G. S. & Mann, J. J. Depression. Lancet 392, 2299–2312 (2018).
Trivedi, M. H. et al. Medication augmentation after the failure of SSRIs for depression. N. Engl. J. Med. 354, 1243–1252 (2006).
Turner, E. H. Esketamine for treatment-resistant depression: seven concerns about efficacy and FDA approval. Lancet Psychiatry 6, 977–979 (2019).
Wilkinson, S. T. et al. The effect of a single dose of intravenous ketamine on suicidal ideation: a systematic review and individual participant data meta-analysis. Am. J. Psychiatry 175, 150–158 (2018).
Yang, Y. et al. Ketamine blocks bursting in the lateral habenula to rapidly relieve depression. Nature 554, 317–322 (2018).
Duman, R. S., Aghajanian, G. K., Sanacora, G. & Krystal, J. H. Synaptic plasticity and depression: new insights from stress and rapid-acting antidepressants. Nat. Med. 22, 238–249 (2016).
Moda-Sava, R. N. et al. Sustained rescue of prefrontal circuit dysfunction by antidepressant-induced spine formation. Science 364, eaat8078 (2019).
Paoletti, P., Bellone, C. & Zhou, Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 14, 383–400 (2013).
Karakas, E. & Furukawa, H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science 344, 992–997 (2014).
Zhu, S. et al. Mechanism of NMDA receptor inhibition and activation. Cell 165, 704–714 (2016).
Jalali-Yazdi, F., Chowdhury, S., Yoshioka, C. & Gouaux, E. Mechanisms for zinc and proton inhibition of the GluN1/GluN2A NMDA receptor. Cell 175, 1520–1532.e15 (2018).
Chou, T. H., Tajima, N., Romero-Hernandez, A. & Furukawa, H. Structural basis of functional transitions in mammalian NMDA receptors. Cell 182, 357–371.e13 (2020).
Jelen, L. A., Young, A. H. & Stone, J. M. Ketamine: a tale of two enantiomers. J. Psychopharmacol. 35, 109–123 (2021).
Erreger, K., Dravid, S. M., Banke, T. G., Wyllie, D. J. & Traynelis, S. F. Subunit-specific gating controls rat NR1/NR2A and NR1/NR2B NMDA channel kinetics and synaptic signalling profiles. J. Physiol. 563, 345–358 (2005).
Burnashev, N. et al. Control by asparagine residues of calcium permeability and magnesium blockade in the NMDA receptor. Science 257, 1415–1419 (1992).
Song, X. et al. Mechanism of NMDA receptor channel block by MK-801 and memantine. Nature 556, 515–519 (2018).
Durham, R. J. et al. Conformational spread and dynamics in allostery of NMDA receptors. Proc. Natl Acad. Sci. USA 117, 3839–3847 (2020).
Iacobucci, G. J. et al. Cross-subunit interactions that stabilize open states mediate gating in NMDA receptors. Proc. Natl Acad. Sci. USA 118, e2007511118 (2021).
Kashiwagi, K. et al. Channel blockers acting at N-methyl-d-aspartate receptors: differential effects of mutations in the vestibule and ion channel pore. Mol. Pharmacol. 61, 533–545 (2002).
Chang, L. et al. Comparison of antidepressant and side effects in mice after intranasal administration of (R,S)-ketamine, (R)-ketamine, and (S)-ketamine. Pharmacol. Biochem. Behav. 181, 53–59 (2019).
Lü, W., Du, J., Goehring, A. & Gouaux, E. Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science 355, eaal3729 (2017).
Zanos, P. et al. NMDAR inhibition-independent antidepressant actions of ketamine metabolites. Nature 533, 481–486 (2016).
Lumsden, E. W. et al. Antidepressant-relevant concentrations of the ketamine metabolite (2R,6R)-hydroxynorketamine do not block NMDA receptor function. Proc. Natl Acad. Sci. USA 116, 5160–5169 (2019).
Lee, C. H. et al. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature 511, 191–197 (2014).
Zhang, J. B. et al. Structural basis of the proton sensitivity of human GluN1-GluN2A NMDA receptors. Cell Rep. 25, 3582–3590.e4 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J., Nakane, T. & Scheres, S. H. W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 (2016).
Schrodinger, LLC Maestro, Version 9.0 (2009).
Vanommeslaeghe, K. & MacKerell, A. D., Jr. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 52, 3144–3154 (2012).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Kumari, R., Kumar, R., Open Source Drug Discovery Consortium & Lynn, A. g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 54, 1951–1962 (2014).
Chen, S. et al. Activation and desensitization mechanism of AMPA receptor-TARP complex by cryo-EM. Cell 170, 1234–1246.e14 (2017).
Meyerson, J. R. et al. Structural basis of kainate subtype glutamate receptor desensitization. Nature 537, 567–571 (2016).
Acknowledgements
We thank B. Zhu, X. Li, C. Liu, F. Meng and Z. Guo for their assistance with cryo-EM data collection; H. Wang, J. Zhang and Y. Gu for assistance with experiments; Y. Sun for advice on human NMDA receptor variants; and M. Poo and E. Xu for proofreading. Financial support is gratefully acknowledged from the National Natural Science Foundation of China (31771115), the National Key R&D Program of China (2017YFA0505700), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB32020000), the Shanghai Municipal Science and Technology Major Project (2018SHZDZX05) and the Thousand Young Talents Program to S.Z.; and the National Natural Science Foundation of China (81625022, 91853205, 81821005) and the Shanghai Municipal Health Commission in China (18431907100 and 19XD1404700) to C.L.
Author information
Authors and Affiliations
Contributions
Y.Z. and T.Z. purified and froze the protein, collected and analysed the cryo-EM data, built the atomic model and conducted electrophysiology on GluN1–GluN2A and GluN1–GluN2B receptors, respectively; F.Y., L.Z., and C.L. carried out the docking and MD simulation; D.D. assisted with binding assays; S.L. and F.G. participated in the data analysis; H.L. provided ketamine compounds; Y.Z., F.Y., T.Z. and S.Z wrote the manuscript with inputs from all the authors. S.Z. conceived the project and supervised the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
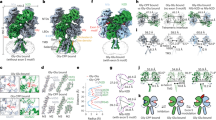
Extended Data Fig. 1 Expression profile and functional validation of human GluN1–GluN2A and GluN1–GluN2B NMDA receptors.
a, Schematic representation of human GluN1EM (grey), GluN2AEM (green) and GluN2BEM (blue) CTD-truncated constructs. b, Representative fluorescence SEC and Coomassie blue gel staining of the purified GluN1–GluN2AEM (left) and GluN1–GluN2BEM receptors (right). Experiments were performed three times independently with similar results. c, d, Representative recording traces (left) and the fitted DRCs (right; mean ± s.d.) for (R,S)-ketamine inhibition on wild-type GluN1–GluN2A and GluN1–GluN2AEM (c), and wild-type GluN1–GluN2B and GluN1–GluN2BEM (d) receptors activated with 100 μM coagonists. Ketamine IC50 values, Hill slopes and numbers are listed in Extended Data Table 3.
Extended Data Fig. 2 Overview of cryo-EM image processing and 3D reconstruction of GluN1–GluN2A and GluN1–GluN2B NMDA receptors.
a, Flowchart of the image processing and 3D reconstruction of human GluN1–GluN2A receptors in complex with S-ketamine, glycine and glutamate. Typical single particles are circled in red from raw micrographs. The 2D class average images show characteristic 2D views in various orientations. The 3D classes with similar conformations were selected and combined through several rounds of 3D classification for final refinement. b, Fourier shell correlation (FSC) curve for the resolution estimation. c, Side view of the cryo-EM density map of GluN1–GluN2A receptor coloured by local resolution estimated by Relion 3.1. d, Pipelines for single-particle analysis and reconstruction of human GluN1–GluN2B receptor in complex with S-ketamine, glycine and glutamate. Same workflow as in a. e, FSC curve for the resolution estimation. f, Side view of the cryo-EM density map of GluN1–GluN2B receptor coloured by local resolution estimated by Relion 3.1.
Extended Data Fig. 3 Representative local densities in GluN1–GluN2A cryo-EM map with fitted atomic model.
a, b, Zoomed-in views of the GluN1-LBD cleft with EM density for glycine (a), and the GluN2A-LBD cleft with glutamate (b). Key binding residues are shown as sticks. c, d, Local densities in extracellular domains of the GluN1 (a) and GluN2A (b) subunits. EM densities are shown as light grey mesh, while the side chains and N-glycosylation sites of residues are represented as sticks.
Extended Data Fig. 4 Functional validation of the ketamine-binding pocket of NMDA receptors.
a, Representative I–V curves for Mg2+ blockage of wild-type GluN1–GluN2A receptors or receptors incorporating a substitution (A, N or V) at GluN2A-L642, recorded in the presence of 100 μM MgCl2. b, c, Plot of 3 μM ketamine inhibition level for wild-type (open circle) and mutant (filled circles) amino acid volume (I, V, A or G) at position GluN2B-L643 or GluN2D-L667, shown with a linear regression. R2 values equal to 0.72 for GluN1–GluN2B and 0.95 for GluN1–GluN2D receptors. d, e, (R,S)-Ketamine DRCs for the GluN1–GluN2A and GluN1–GluN2B wild-type or mutant receptors incorporating a substitution (A, L or T) at GluN1-V644. All IC50 values, Hill slopes and numbers of oocytes are listed in Extended Data Table 3. f, Schematic representation of the S-ketamine binding site on GluN1–GluN2B receptors analysed by Ligplot+.
Extended Data Fig. 5 Molecular basis of S- and R-ketamine-induced inhibition of GluN1–GluN2A receptors.
a, Chemical structures of left-handed S-ketamine and right-handed R-ketamine. b, r.m.s.d. trajectories for each chain (excluding M1–M2 loops) and R-ketamine were calculated on Cα atoms based on the initial structure within the whole simulation time of 500 ns. c, Left, R-ketamine poses obtained in MD simulation along the whole simulation time. Right, schematic diagram of R-ketamine and TMD interactions at 500 ns snapshot extracted from MD simulation. Residues involved in the hydrophobic interactions are shown as starbursts. d, Inhibition by 2 μM S-ketamine of NMDA receptor activity induced by saturating agonists in wild-type GluN1–GluN2A (0.606 ± 0.018, n = 9 oocytes), GluN1(N616A)–GluN2A (0.020 ± 0.004, n = 3), GluN1(N616Q)–GluN2A (0.027 ± 0.006, n = 3), GluN1–GluN2A(L642A) (0.041 ± 0.012, n = 3), GluN1–GluN2A(L642V) (0.152 ± 0.007, n = 3), GluN1–GluN2A(L642N) (0.020 ± 0.001, n = 3), GluN1(V644A)–GluN2A (0.333 ± 0.038, n = 3), GluN1(V644L)–GluN2A (0.455 ± 0.012, n = 3), GluN1(V644T)–GluN2A (0.602 ± 0.020, n = 3), GluN1(T648V)–GluN2A (0.621 ± 0.015, n = 3), GluN1–GluN2A(T646V) (0.532 ± 0.024, n = 4) and GluN1–GluN2A(N615A) (0.597 ± 0.008, n = 3) receptors. e, Inhibition by 2 μM R-ketamine of wild-type GluN1–GluN2A (0.404 ± 0.017, n = 9), GluN1(N616A)–GluN2A (0.008 ± 0.001, n = 3), GluN1(N616Q)–GluN2A (0.049 ± 0.008, n = 3), GluN1–GluN2A(L642A) (0.014 ± 0.001, n = 3), GluN1–GluN2A(L642V) (0.065 ± 0.009, n = 3), GluN1–GluN2A(L642N) (0.019 ± 0.001, n = 3), GluN1(T648V)–GluN2A (0.330 ± 0.021, n = 3), GluN1–GluN2A(T646V) (0.423 ± 0.021, n = 4) and GluN1–GluN2A(N615A) (0.344 ± 0.01, n = 3) receptors. f, Superposition of the 500 ns snapshots extracted from MD simulation of S-ketamine (pink) and R-ketamine (cyan) systems, respectively. g, Inhibition by 2 μM R- and S-ketamine of GluN1–GluN2A(N614A) (0.138 ± 0.009, 0.509 ± 0.014, n = 3) and GluN1–GluN2A(N614Q) (0.140 ± 0.006, 0.633 ± 0.014, n = 3) receptors. In d, e, g, all data shown are mean ± s.e.m.; P values are determined by one-way ANOVA with Dunnett’s multiple comparison test (****P < 0.0001). Each data point represents the result of one oocyte. h, R-ketamine (left) and S-ketamine (right) DRCs (mean ± s.d.) for wild-type GluN1–GluN2A (IC50: 2.39 ± 0.45 μM, n = 4 oocytes; 0.60 ± 0.03 μM, n = 3), GluN1–GluN2A(N614A) (13.66 ± 2.49 μM, n = 4; 1.22 ± 0.44 μM, n = 4) and GluN1–GluN2A(N614Q) (39.94 ± 6.72 μM, n = 4; 1.41 ± 0.38 μM, n = 4) receptors.
Extended Data Fig. 6 Sequence alignment and structural comparison of TMD in ionotropic GluRs.
a, Sequence alignment of TM2–TM3 segments in human GluN1, GluN2A, GluN2B, GluN2C, GluN2D, GluA2 and GluK2, Xenopus GluN1, GluN2B and rat GluA2 subunits. The critical residues involved in ketamine binding are highlighted in yellow, and their homologous sites in ionotropic GluRs are marked in rectangles. b, Superposition of the TM2 and TM3 segments between the S-ketamine (in brick red)-bound GluN1–GluN2BEM receptor and the MK-801 (in red)-bound GluN1–GluN2B(ΔNTD) receptor (PDB code: 5UN1)20. c, MK-801-bound TMD of the triheteromeric GluN1–GluN2A–GluN2B NMDA receptor, viewed parallel to the membrane (PDB code: 5UOW)25. MK-801 binding residues were analysed by LigPlot+ (right). d, e, Superposition of the TM2 and TM3 segments between the S-ketamine bound GluN1–GluN2AEM receptor and GluA2 AMPA receptor (PDB code: 5VOT)41 (d) or the GluK2 kainate receptor (PDB code: 5KUF)42 (e). All superpositions are based on the α-carbon atoms of the conserved SYTANL region.
Supplementary information
Supplementary Figure 1
Uncropped gels presented in the Extended Data Fig. 1.
Video 1
Motions of GluN1-GluN2A TMD in complex with S-ketamine (brick-red) and R-ketamine (blue) during the 500 ns MD simulation. The TMD of GluN1 and GluN2A subunits are shown in grey and green, respectively.
Rights and permissions
About this article
Cite this article
Zhang, Y., Ye, F., Zhang, T. et al. Structural basis of ketamine action on human NMDA receptors. Nature 596, 301–305 (2021). https://doi.org/10.1038/s41586-021-03769-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03769-9
This article is cited by
-
Safety and efficacy of low-dose esketamine in laparoscopic cholecystectomy: a prospective, double-blind randomized controlled trial
BMC Anesthesiology (2024)
-
Features of biliary tract diseases in ketamine abusers: a systematic review of case reports
Journal of Medical Case Reports (2024)
-
Major depressive disorder: hypothesis, mechanism, prevention and treatment
Signal Transduction and Targeted Therapy (2024)
-
Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits
Nature Structural & Molecular Biology (2023)
-
Structural and functional imaging of brains
Science China Chemistry (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.