Abstract
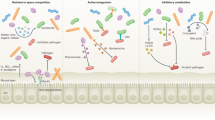
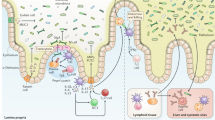
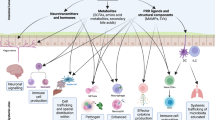
The microbiome has an important role in human health. Changes in the microbiota can confer resistance to or promote infection by pathogenic bacteria. Antibiotics have a profound impact on the microbiota that alters the nutritional landscape of the gut and can lead to the expansion of pathogenic populations. Pathogenic bacteria exploit microbiota-derived sources of carbon and nitrogen as nutrients and regulatory signals to promote their own growth and virulence. By eliciting inflammation, these bacteria alter the intestinal environment and use unique systems for respiration and metal acquisition to drive their expansion. Unravelling the interactions between the microbiota, the host and pathogenic bacteria will produce strategies for manipulating the microbiota against infectious diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007).
The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Ley, R. E., Peterson, D. A. & Gordon, J. I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 (2006).
Yurist-Doutsch, S., Arrieta, M. C., Vogt, S. L. & Finlay, B. B. Gastrointestinal microbiota-mediated control of enteric pathogens. Annu. Rev. Genet. 48, 361–382 (2014).
Sassone-Corsi, M. & Raffatellu, M. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J. Immunol. 194, 4081–4087 (2015).
Cameron, E. A. & Sperandio, V. Frenemies: signaling and nutritional integration in pathogen–microbiota–host interactions. Cell Host Microbe 18, 275–284 (2015).
Pacheco, A. R. & Sperandio, V. Enteric pathogens exploit the microbiota-generated nutritional environment of the gut. Microbiol. Spectr. 3, MBP-0001-2014 (2015).
Bohnhoff, M., Drake, B. L. & Miller, C. P. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc. Soc. Exp. Biol. Med. 86, 132–137 (1954).
Ferreira, R. B. et al. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS ONE 6, e20338 (2011).
Sprinz, H. et al. The response of the germfree guinea pig to oral bacterial challenge with Escherichia coli and Shigella flexneri. Am. J. Pathol. 39, 681–695 (1961).
Zachar, Z. & Savage, D. C. Microbial interference and colonization of the murine gastrointestinal tract by Listeria monocytogenes. Infect. Immun. 23, 168–174 (1979).
Kamada, N. et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329 (2012). This study showed that enteric pathogens use virulence genes to compete with the microbiota for nutrients in the gut.
Ghosh, S. et al. Colonic microbiota alters host susceptibility to infectious colitis by modulating inflammation, redox status, and ion transporter gene expression. Am. J. Physiol. Gastrointest. Liver Physiol. 301, G39–G49 (2011).
Willing, B. P., Vacharaksa, A., Croxen, M., Thanachayanont, T. & Finlay, B. B. Altering host resistance to infections through microbial transplantation. PLoS ONE 6, e26988 (2011).
Kampmann, C., Dicksved, J., Engstrand, L. & Rautelin, H. Composition of human faecal microbiota in resistance to Campylobacter infection. Clin. Microbiol. Infect. 22, 61.e1–61.e8 (2016).
Kau, A. L., Ahern, P. P., Griffin, N. W., Goodman, A. L. & Gordon, J. I. Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 (2011).
Zumbrun, S. D. et al. Dietary choice affects Shiga toxin-producing Escherichia coli (STEC) O157:H7 colonization and disease. Proc. Natl Acad. Sci. USA 110, E2126–E2133 (2013).
Wlodarska, M., Willing, B. P., Bravo, D. M. & Finlay, B. B. Phytonutrient diet supplementation promotes beneficial Clostridia species and intestinal mucus secretion resulting in protection against enteric infection. Sci. Rep. 5, 9253 (2015).
Modi, S. R., Collins, J. J. & Relman, D. A. Antibiotics and the gut microbiota. J. Clin. Invest. 124, 4212–4218 (2014).
Leffler, D. A. & Lamont, J. T. Clostridium difficile infection. N. Engl. J. Med. 373, 287–288 (2015).
Pavia, A. T. et al. Epidemiologic evidence that prior antimicrobial exposure decreases resistance to infection by antimicrobial-sensitive Salmonella. J. Infect. Dis. 161, 255–260 (1990).
Ng, K. M. et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99 (2013). This study showed that treatment with antibiotics enhances the abundance of host sialic acid that can be harvested by the microbiota, which promotes the expansion of enteric pathogens.
Ferreyra, J. A. et al. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 16, 770–777 (2014).
Ferreyra, J. A., Ng, K. M. & Sonnenburg, J. L. The enteric two-step: nutritional strategies of bacterial pathogens within the gut. Cell. Microbiol. 16, 993–1003 (2014).
Rakoff-Nahoum, S., Coyne, M. J. & Comstock, L. E. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr. Biol. 24, 40–49 (2014).
Alverdy, J., Chi, H. S. & Sheldon, G. F. The effect of parenteral nutrition on gastrointestinal immunity. The importance of enteral stimulation. Ann. Surg. 202, 681–684 (1985).
Fischbach, M. A. & Sonnenburg, J. L. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10, 336–347 (2011).
Chow, W. L. & Lee, Y. K. Free fucose is a danger signal to human intestinal epithelial cells. Br. J. Nutr. 99, 449–454 (2008).
Bourlioux, P., Koletzko, B., Guarner, F. & Braesco, V. The intestine and its microflora are partners for the protection of the host: report on the Danone Symposium “The Intelligent Intestine,” held in Paris, June 14, 2002. Am. J. Clin. Nutr. 78, 675–683 (2003).
Bry, L., Falk, P. G., Midtvedt, T. & Gordon, J. I. A model of host–microbial interactions in an open mammalian ecosystem. Science 273, 1380–1383 (1996).
Hooper, L. V., Xu, J., Falk, P. G., Midtvedt, T. & Gordon, J. I. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl Acad. Sci. USA 96, 9833–9838 (1999).
Fabich, A. J. et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 76, 1143–1152 (2008).
Autieri, S. M. et al. L-fucose stimulates utilization of D-ribose by Escherichia coli MG1655 ΔfucAO and E. coli Nissle 1917 ΔfucAO mutants in the mouse intestine and in M9 minimal medium. Infect. Immun. 75, 5465–5475 (2007).
Pacheco, A. R. et al. Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–117 (2012). This paper showed that sugars from the mucus that are released by the microbiota can be perceived as signals by enteric pathogens to regulate the expression of virulence genes.
Schauer, D. B. & Falkow, S. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61, 2486–2492 (1993).
Szabady, R. L., Lokuta, M. A., Walters, K. B., Huttenlocher, A. & Welch, R. A. Modulation of neutrophil function by a secreted mucinase of Escherichia coli O157:H7. PLoS Pathog. 5, e1000320 (2009).
Curtis, M. M. et al. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16, 759–769 (2014). This study revealed that metabolites that are produced by the microbiota can be exploited as cues to increase the virulence of enteric pathogens.
Macy, J. M., Ljungdahl, L. G. & Gottschalk, G. Pathway of succinate and propionate formation in Bacteroides fragilis. J. Bacteriol. 134, 84–91 (1978).
Carter, P. B. & Collins, F. M. The route of enteric infection in normal mice. J. Exp. Med. 139, 1189–1203 (1974).
Lawhon, S. D., Maurer, R., Suyemoto, M. & Altier, C. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46, 1451–1464 (2002).
Takao, M., Yen, H. & Tobe, T. LeuO enhances butyrate-induced virulence expression through a positive regulatory loop in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 93, 1302–1313 (2014).
Karmali, M. A., Petric, M., Lim, C., Fleming, P. C. & Steele, B. T. Escherichia coli cytotoxin, haemolytic-uraemic syndrome, and haemorrhagic colitis. Lancet 2, 1299–1300 (1983).
Fukuda, S. et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547 (2011). This study showed that the acetate that is produced by certain probiotic bacteria can enhance the barrier function of the gut epithelium to protect the host from enteric infections.
Bertin, Y. et al. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ. Microbiol. 13, 365–377 (2011).
Garsin, D. A. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nature Rev. Microbiol. 8, 290–295 (2010).
Korbel, J. O. et al. Systematic association of genes to phenotypes by genome and literature mining. PLoS Biol. 3, e134 (2005).
Thiennimitr, P. et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl Acad. Sci. USA 108, 17480–17485 (2011).
Joseph, B. et al. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188, 556–568 (2006).
Kendall, M. M., Gruber, C. C., Parker, C. T. & Sperandio, V. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3, 00050-12 (2012).
Anderson, C. J., Clark, D. E., Adli, M. & Kendall, M. M. Ethanolamine signaling promotes Salmonella niche recognition and adaptation during infection. PLoS Pathog. 11, e1005278 (2015).
Maier, L. et al. Microbiota-derived hydrogen fuels Salmonella Typhimurium invasion of the gut ecosystem. Cell Host Microbe 14, 641–651 (2013).
Maltby, R., Leatham-Jensen, M. P., Gibson, T., Cohen, P. S. & Conway, T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS ONE 8, e53957 (2013).
Fabich, A. J. et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 76, 1143–1152 (2008).
Hsiao, A. et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515, 423–426 (2014).
Hughes, D. T. et al. Chemical sensing in mammalian host–bacterial commensal associations. Proc. Natl Acad. Sci. USA 107, 9831–9836 (2010).
Bachmann, V. et al. Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Negl. Trop. Dis. 9, e0004031 (2015).
Hörger, S., Schultheiss, G. & Diener, M. Segment-specific effects of epinephrine on ion transport in the colon of the rat. Am. J. Physiol. 275, G1367–G1376 (1998).
Yano, J. M. et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276 (2015).
Asano, Y. et al. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G1288–G1295 (2012).
Furness, J. B. Types of neurons in the enteric nervous system. J. Auton. Nerv. Syst. 81, 87–96 (2000).
Curtis, M. M. & Sperandio, V. A complex relationship: the interaction among symbiotic microbes, invading pathogens, and their mammalian host. Mucosal Immunol. 4, 133–138 (2011).
Moreira, C. G., Weinshenker, D. & Sperandio, V. QseC mediates Salmonella enterica serovar Typhimurium virulence in vitro and in vivo. Infect. Immun. 78, 914–926 (2010).
Sperandio, V., Torres, A. G., Jarvis, B., Nataro, J. P. & Kaper, J. B. Bacteria–host communication: the language of hormones. Proc. Natl Acad. Sci. USA 100, 8951–8956 (2003). This paper describes how signals from the microbiota and host converge to enhance virulence in enteric pathogens.
Nakano, M., Takahashi, A., Sakai, Y. & Nakaya, Y. Modulation of pathogenicity with norepinephrine related to the type III secretion system of Vibrio parahaemolyticus. J. Infect. Dis. 195, 1353–1360 (2007).
Clarke, M. B., Hughes, D. T., Zhu, C., Boedeker, E. C. & Sperandio, V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl Acad. Sci. USA 103, 10420–10425 (2006).
Reading, N. C., Rasko, D. A., Torres, A. G. & Sperandio, V. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl Acad. Sci. USA 106, 5889–5894 (2009).
Seksik, P. et al. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut 52, 237–242 (2003).
Gophna, U., Sommerfeld, K., Gophna, S., Doolittle, W. F. & Veldhuyzen van Zanten, S. J. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J. Clin. Microbiol. 44, 4136–4141 (2006).
Baumgart, M. et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 1, 403–418 (2007).
Walker, A. W. et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 11, 7 (2011).
Gevers, D. et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15, 382–392 (2014).
Chiodini, R. J. et al. Microbial population differentials between mucosal and submucosal intestinal tissues in advanced Crohn's disease of the ileum. PLoS ONE 10, e0134382 (2015).
Lupp, C. et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129; corrigendum 2, 204 (2007). This paper reported that the induction of inflammation in the host disrupts the microbiota and promotes a bloom of Enterobacteriaceae.
Garrett, W. S. et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8, 292–300 (2010).
Raffatellu, M. et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nature Med. 14, 421–428 (2008).
Godinez, I. et al. T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect. Immun. 76, 2008–2017 (2008).
Pickard, J. M. et al. Rapid fucosylation of intestinal epithelium sustains host–commensal symbiosis in sickness. Nature 514, 638–641 (2014). This paper showed that cytokines that are induced by enteric infection promote fucosylation in the host.
Pham, T. A. et al. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe 16, 504–516 (2014).
Sonnenburg, J. L. et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959 (2005).
Ansong, C. et al. Studying Salmonellae and Yersiniae host–pathogen interactions using integrated 'omics and modeling. Curr. Top. Microbiol. Immunol. 363, 21–41 (2013).
Harper, R. W. et al. Differential regulation of dual NADPH oxidases/peroxidases, Duox1 and Duox2, by Th1 and Th2 cytokines in respiratory tract epithelium. FEBS Lett. 579, 4911–4917 (2005).
Haberman, Y. et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Invest. 124, 3617–3633 (2014).
Salzman, A. et al. Induction and activity of nitric oxide synthase in cultured human intestinal epithelial monolayers. Am. J. Physiol. 270, G565–G573 (1996).
Palmer, R. M., Rees, D. D., Ashton, D. S. & Moncada, S. L-arginine is the physiological precursor for the formation of nitric oxide in endothelium-dependent relaxation. Biochem. Biophys. Res. Commun. 153, 1251–1256 (1988).
Lundberg, J. O., Weitzberg, E., Lundberg, J. M. & Alving, K. Intragastric nitric oxide production in humans: measurements in expelled air. Gut 35, 1543–1546 (1994).
Singer, I. I. et al. Expression of inducible nitric oxide synthase and nitrotyrosine in colonic epithelium in inflammatory bowel disease. Gastroenterology 111, 871–885 (1996).
Enocksson, A., Lundberg, J., Weitzberg, E., Norrby-Teglund, A. & Svenungsson, B. Rectal nitric oxide gas and stool cytokine levels during the course of infectious gastroenteritis. Clin. Diagn. Lab. Immunol. 11, 250–254 (2004).
Bai, P. et al. Protein tyrosine nitration and poly(ADP-ribose) polymerase activation in N-methyl-N-nitro-N-nitrosoguanidine-treated thymocytes: implication for cytotoxicity. Toxicol. Lett. 170, 203–213 (2007).
Dudhgaonkar, S. P., Tandan, S. K., Kumar, D., Raviprakash, V. & Kataria, M. Influence of simultaneous inhibition of cyclooxygenase-2 and inducible nitric oxide synthase in experimental colitis in rats. Inflammopharmacology 15, 188–195 (2007).
Winter, S. E. et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013). This study revealed that nitrate respiration drives the expansion of commensal E. coli during colitis.
Winter, S. E. & Baumler, A. J. Why related bacterial species bloom simultaneously in the gut: principles underlying the 'like will to like' concept. Cell. Microbiol. 16, 179–184 (2014).
Rivera-Chávez, F. & Baumler, A. J. The pyromaniac inside you: Salmonella metabolism in the host gut. Annu. Rev. Microbiol. 69, 31–48 (2015).
Galán, J. E. & Curtiss, R., III. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl Acad. Sci. USA 86, 6383–6387 (1989).
Hensel, M. et al. Simultaneous identification of bacterial virulence genes by negative selection. Science 269, 400–403 (1995).
Tsolis, R. M., Adams, L. G., Ficht, T. A. & Baumler, A. J. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67, 4879–4885 (1999).
Barthel, M. et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71, 2839–2858 (2003).
Coburn, B., Li, Y., Owen, D., Vallance, B. A. & Finlay, B. B. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect. Immun. 73, 3219–3227 (2005).
Stecher, B. et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189 (2007).
Lawley, T. D. et al. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect. Immun. 76, 403–416 (2008).
Lopez, C. A. et al. Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. mBio 3, e00143-12 (2012).
Lopez, C. A., Rivera-Chavez, F., Byndloss, M. X. & Baumler, A. J. The periplasmic nitrate reductase NapABC supports luminal growth of Salmonella enterica serovar Typhimurium during colitis. Infect. Immun. 83, 3470–3478 (2015).
Levitt, M. D., Furne, J., Springfield, J., Suarez, F. & DeMaster, E. Detoxification of hydrogen sulfide and methanethiol in the cecal mucosa. J. Clin. Invest. 104, 1107–1114 (1999).
Harris, J. C., Dupont, H. L. & Hornick, R. B. Fecal leukocytes in diarrheal illness. Ann. Intern. Med. 76, 697–703 (1972).
Loetscher, Y. et al. Salmonella transiently reside in luminal neutrophils in the inflamed gut. PLoS ONE 7, e34812 (2012).
Winter, S. E. et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010). This paper showed that tetrathionate is a unique electron acceptor that is induced through inflammation that promotes expansion of S . Typhimurium.
Muller, H. J. Partial list of biological institutes and biologists doing experimental work in Russia at the present time. Science 57, 472–473 (1923).
Raffatellu, M. et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5, 476–486 (2009).
Behnsen, J. et al. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 40, 262–273 (2014).
Goetz, D. H. et al. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 10, 1033–1043 (2002).
Flo, T. H. et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921 (2004).
Deriu, E. et al. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14, 26–37 (2013). This paper revealed that probiotic strains of E. coli restrict infection of the gut with S . Typhimurium by competing for sources of iron.
Cascales, E. et al. Colicin biology. Microbiol. Mol. Biol. Rev. 71, 158–229 (2007).
Guterman, S. K. Colicin B: mode of action and inhibition by enterochelin. J. Bacteriol. 114, 1217–1224 (1973).
Cardelli, J. & Konisky, J. Isolation and characterization of an Escherichia coli mutant tolerant to colicins Ia and Ib. J. Bacteriol. 119, 379–385 (1974).
Patzer, S. I., Baquero, M. R., Bravo, D., Moreno, F. & Hantke, K. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149, 2557–2570 (2003).
Hooper, L. V. et al. Molecular analysis of commensal host–microbial relationships in the intestine. Science 291, 881–884 (2001). This was the first report to show that the microbiota can change the expression of mammalian genes to modulate the immune system of the host.
Wu, M. et al. Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science 350, aac5992 (2015).
Earle, K. A. et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18, 478–488 (2015).
Bouslimani, A. et al. Molecular cartography of the human skin surface in 3D. Proc. Natl Acad. Sci. USA 112, E2120–E2129 (2015).
Marcobal, A. et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 7, 1933–1943 (2013).
Lau, W., Fischbach, M. A., Osbourn, A. & Sattely, E. S. Key applications of plant metabolic engineering. PLoS Biol. 12, e1001879 (2014).
Marcobal, A. et al. Metabolome progression during early gut microbial colonization of gnotobiotic mice. Sci. Rep. 5, 11589 (2015).
Rath, C. M. et al. Molecular analysis of model gut microbiotas by imaging mass spectrometry and nanodesorption electrospray ionization reveals dietary metabolite transformations. Anal. Chem. 84, 9259–9267 (2012). This study used imaging mass spectrometry to map differences in the gut metabolic landscape that are promoted by the microbiota.
Dorrestein, P. C., Mazmanian, S. K. & Knight, R. Finding the missing links among metabolites, microbes, and the host. Immunity 40, 824–832 (2014).
Antunes, L. C. et al. Antivirulence activity of the human gut metabolome. mBio 5, e01183-14 (2014).
Antunes, L. C. et al. Impact of Salmonella infection on host hormone metabolism revealed by metabolomics. Infect. Immun. 79, 1759–1769 (2011).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015). This paper showed that the introduction of a single species of bacteria that can produce secondary bile salts confers the host with resistance to the expansion of C. difficile after treatment with antibiotics.
Koenigsknecht, M. J. & Young, V. B. Faecal microbiota transplantation for the treatment of recurrent Clostridium difficile infection: current promise and future needs. Curr. Opin. Gastroenterol. 29, 628–632 (2013).
Sekirov, I. et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76, 4726–4736 (2008).
Louis, E., Libioulle, C., Reenaers, C., Belaiche, J. & Georges, M. Genomics of inflammatory bowel diseases: basis for a new molecular classification and new therapeutic strategies of these diseases [in French]. Rev. Med. Liege 64 S1, 24–28 (2009).
Meynell, G. G. Antibacterial mechanisms of the mouse gut. II. The role of Eh and volatile fatty acids in the normal gut. Br. J. Exp. Pathol. 44, 209–219 (1963).
Donohoe, D. R., Wali, A., Brylawski, B. P. & Bultman, S. J. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PLoS ONE 7, e46589 (2012).
Atarashi, K., Umesaki, Y. & Honda, K. Microbiotal influence on T cell subset development. Semin. Immunol. 23, 146–153 (2011).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Spees, A. M. et al. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. mBio 4, e00430-13 (2013).
Itoh, K. & Freter, R. Control of Escherichia coli populations by a combination of indigenous clostridia and lactobacilli in gnotobiotic mice and continuous-flow cultures. Infect. Immun. 57, 559–565 (1989).
Wells, J. E. & Hylemon, P. B. Identification and characterization of a bile acid 7α-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7α-dehydroxylating strain isolated from human feces. Appl. Environ. Microbiol. 66, 1107–1113 (2000).
Wilson, K. H. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J. Clin. Microbiol. 18, 1017–1019 (1983).
Sorg, J. A. & Sonenshein, A. L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190, 2505–2512 (2008).
Theriot, C. M. et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nature Commun. 5, 3114 (2014).
Weingarden, A. R. et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Physiol. Gastrointest. Liver Physiol. 306, G310–G319 (2014).
Bernstein, H., Bernstein, C., Payne, C. M., Dvorakova, K. & Garewal, H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res. 589, 47–65 (2005).
Acknowledgements
Work in the V.S. laboratory is supported by US National Institutes of Health (NIH) grants AI053067, AI077613, AI05135 and AI114511. Work in the A.J.B. laboratory is supported by US Department of Agriculture grant 2015-67015-22930 and NIH grants AI044170, AI096528, AI112445, AI114922 and AI117940. The contents of this Review are solely the responsibility of the authors and do not necessarily represent the official views of the NIH National Institute of Allergy and Infectious Diseases.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available at www.nature.com.reprints.
Rights and permissions
About this article
Cite this article
Bäumler, A., Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535, 85–93 (2016). https://doi.org/10.1038/nature18849
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18849
This article is cited by
-
Bacterial community and diversity in the rumen of 11 Mongolian cattle as revealed by 16S rRNA amplicon sequencing
Scientific Reports (2024)
-
Alterations in the gut microbiota and their metabolites in human intestinal epithelial cells of patients with colorectal cancer
Molecular Biology Reports (2024)
-
Dietary Macleaya cordata extract supplementation improves the growth performance and gut health of broiler chickens with necrotic enteritis
Journal of Animal Science and Biotechnology (2023)
-
Multi-omics analysis of fecal samples in colorectal cancer Egyptians patients: a pilot study
BMC Microbiology (2023)
-
Aspirin inhibits rotavirus replication and alters rat gut microbial composition
Virology Journal (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.