Abstract
Lymphoid organs, in which antigen presenting cells (APCs) are in close proximity to T cells, are the ideal microenvironment for efficient priming and amplification of T-cell responses1. However, the systemic delivery of vaccine antigens into dendritic cells (DCs) is hampered by various technical challenges. Here we show that DCs can be targeted precisely and effectively in vivo using intravenously administered RNA-lipoplexes (RNA-LPX) based on well-known lipid carriers by optimally adjusting net charge, without the need for functionalization of particles with molecular ligands. The LPX protects RNA from extracellular ribonucleases and mediates its efficient uptake and expression of the encoded antigen by DC populations and macrophages in various lymphoid compartments. RNA-LPX triggers interferon-α (IFNα) release by plasmacytoid DCs and macrophages. Consequently, DC maturation in situ and inflammatory immune mechanisms reminiscent of those in the early systemic phase of viral infection are activated2. We show that RNA-LPX encoding viral or mutant neo-antigens or endogenous self-antigens induce strong effector and memory T-cell responses, and mediate potent IFNα-dependent rejection of progressive tumours. A phase I dose-escalation trial testing RNA-LPX that encode shared tumour antigens is ongoing. In the first three melanoma patients treated at a low-dose level, IFNα and strong antigen-specific T-cell responses were induced, supporting the identified mode of action and potency. As any polypeptide-based antigen can be encoded as RNA3,4, RNA-LPX represent a universally applicable vaccine class for systemic DC targeting and synchronized induction of both highly potent adaptive as well as type-I-IFN-mediated innate immune mechanisms for cancer immunotherapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
07 June 2016
The competing financial interests statement did not display correctly online when this paper was first published; this has been corrected and the statement is now available.
References
Zinkernagel, R. M. et al. Antigen localisation regulates immune responses in a dose- and time-dependent fashion: a geographical view of immune reactivity. Immunol. Rev. 156, 199–209 (1997).
Stetson, D. B. & Medzhitov, R. Type I interferons in host defense. Immunity 25, 373–381 (2006).
Boczkowski, D., Nair, S. K., Snyder, D. & Gilboa, E. Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J. Exp. Med. 184, 465–472 (1996).
Sahin, U., Karikó, K. & Türeci, Ö. mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discov. 13, 759–780 (2014).
Banchereau, J. & Steinman, R. M. Dendritic cells and the control of immunity. Nature 392, 245–252 (1998).
Tacken, P. J., de Vries, I. J. M., Torensma, R. & Figdor, C. G. Dendritic-cell immunotherapy: from ex vivo loading to in vivo targeting. Nat. Rev. Immunol. 7, 790–802 (2007).
Phua, K. K. L. Towards targeted delivery systems: ligand conjugation strategies for mRNA nanoparticle tumor vaccines. J. Immunol. Res. 680620 (2015).
Mitragotri, S., Burke, P. A. & Langer, R. Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies. Nat. Rev. Drug Discov. 13, 655–672 (2014).
Brito, L. A. et al. A cationic nanoemulsion for the delivery of next generation RNA vaccines. Mol. Ther. 22, 2118–2129 (2014).
Pollard, C. et al. Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol. Ther. 21, 251–259 (2013).
Zhou, W. Z. et al. RNA melanoma vaccine: induction of antitumor immunity by human glycoprotein 100 mRNA immunization. Hum. Gene Ther. 10, 2719–2724 (1999).
Hess, P. R., Boczkowski, D., Nair, S. K., Snyder, D. & Gilboa, E. Vaccination with mRNAs encoding tumor-associated antigens and granulocyte-macrophage colony-stimulating factor efficiently primes CTL responses, but is insufficient to overcome tolerance to a model tumor/self antigen. Cancer Immunol. Immunother. 55, 672–683 (2006).
Perche, F. et al. Enhancement of dendritic cells transfection in vivo and of vaccination against B16F10 melanoma with mannosylated histidylated lipopolyplexes loaded with tumor antigen messenger RNA. Nanomedicine 7, 445–453 (2011).
Blanco, E., Shen, H. & Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 33, 941–951 (2015).
Lee, E. R. et al. Detailed analysis of structures and formulations of cationic lipids for efficient gene transfer to the lung. Hum. Gene Ther. 7, 1701–1717 (1996).
Liu, Y. et al. Factors influencing the efficiency of cationic liposome-mediated intravenous gene delivery. Nat. Biotechnol. 15, 167–173 (1997).
Diken, M. et al. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 18, 702–708 (2011).
Sallusto, F., Cella, M., Danieli, C. & Lanzavecchia, A. Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products. J. Exp. Med. 182, 389–400 (1995).
Platt, C. D. et al. Mature dendritic cells use endocytic receptors to capture and present antigens. Proc. Natl Acad. Sci. USA 107, 4287–4292 (2010).
Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S. & Reis e Sousa & C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531 (2004).
Kreiter, S. et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520, 692–696 (2015).
Le Bon, A. et al. Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat. Immunol. 4, 1009–1015 (2003).
Bacher, N. et al. Interferon-α suppresses cAMP to disarm human regulatory T cells. Cancer Res. 73, 5647–5656 (2013).
Zitvogel, L., Galluzzi, L., Kepp, O., Smyth, M. J. & Kroemer, G. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 15, 405–414 (2015).
Janeway, C. A. Jr The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol. Today 13, 11–16 (1992).
Janeway, C. A. Jr & Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 (2002).
Swiecki, M., Gilfillan, S., Vermi, W., Wang, Y. & Colonna, M. Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8+ T cell accrual. Immunity 33, 955–966 (2010).
Kamphuis, E., Junt, T., Waibler, Z., Forster, R. & Kalinke, U. Type I interferons directly regulate lymphocyte recirculation and cause transient blood lymphopenia. Blood 108, 3253–3261 (2006).
Müller, U. et al. Functional role of type I and type II interferons in antiviral defense. Science 264, 1918–1921 (1994).
Jung, S. et al. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity 17, 211–220 (2002).
Huang, A. Y. et al. The immunodominant major histocompatibility complex class I-restricted antigen of a murine colon tumor derives from an endogenous retroviral gene product. Proc. Natl Acad. Sci. USA 93, 9730–9735 (1996).
Lin, K.-Y. et al. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 21–26 (1996).
Chuang, C.-M., Monie, A., Wu, A. & Hung, C.-F. Combination of apigenin treatment with therapeutic HPV DNA vaccination generates enhanced therapeutic antitumor effects. J. Biomed. Sci. 16, 49 (2009).
Holtkamp, S. et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 108, 4009–4017 (2006).
Kreiter, S. et al. Increased antigen presentation efficiency by coupling antigens to MHC class I trafficking signals. J. Immunol. 180, 309–318 (2008).
Kuhn, A. N. et al. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 17, 961–971 (2010).
Slansky, J. E. et al. Enhanced antigen-specific antitumor immunity with altered peptide ligands that stabilize the MHC-peptide-TCR complex. Immunity 13, 529–538 (2000).
Kenter, G. G. et al. Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. N. Engl. J. Med. 361, 1838–1847 (2009).
Kreiter, S. et al. Simultaneous ex vivo quantification of antigen-specific CD4+ and CD8+ T cell responses using in vitro transcribed RNA. Cancer Immunol. Immunother. 56, 1577–1587 (2007).
Bangham, A. D., Hill, M. W. & Miller, N. G. A. in Methods in Membrane Biology (ed. Korn, E. D. ) 1–68 (Springer US, 1974).
Duzgunes, N. in Methods in Enzymology (Academic Press Inc, 2009).
Batzri, S. & Korn, E. D. Single bilayer liposomes prepared without sonication. Biochim. Biophys. Acta 298, 1015–1019 (1973).
Barichello, J. M., Ishida, T. & Kiwada, H. Complexation of siRNA and pDNA with cationic liposomes: the important aspects in lipoplex preparation. Methods Mol. Biol. 605, 461–472 (2010).
Racoosin, E. L. & Swanson, J. A. Macrophage colony-stimulating factor (rM-CSF) stimulates pinocytosis in bone marrow-derived macrophages. J. Exp. Med. 170, 1635–1648 (1989).
Sarkar, K., Kruhlak, M. J., Erlandsen, S. L. & Shaw, S. Selective inhibition by rottlerin of macropinocytosis in monocyte-derived dendritic cells. Immunology 116, 513–524 (2005).
Bar-On, L. & Jung, S. Defining in vivo dendritic cell functions using CD11c-DTR transgenic mice. Methods Mol. Biol. 595, 429–442 (2010).
Aichele, P. et al. Macrophages of the splenic marginal zone are essential for trapping of blood-borne particulate antigen but dispensable for induction of specific T cell responses. J. Immunol. 171, 1148–1155 (2003).
Wexler, H. Accurate identification of experimental pulmonary metastases. J. Natl. Cancer Inst. 36, 641–645 (1966).
Chen, Y. T. et al. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc. Natl Acad. Sci. USA 94, 1914–1918 (1997).
Brichard, V. et al. The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J. Exp. Med. 178, 489–495 (1993).
Gaugler, B. et al. Human gene MAGE-3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J. Exp. Med. 179, 921–930 (1994).
Simon, P. et al. Functional TCR retrieval from single antigen-specific human T cells reveals multiple novel epitopes. Cancer Immunol. Res. 2, 1230–1244 (2014).
Acknowledgements
The authors thank M. Holzmann, R. Roth, U. Schmitt, M. Brkic, A. König, C. Worm, N. Krimmel, A.-K. Thiel, C. Bender, M. Suchan, A.-L. Popa, P. Bezerra Gomes, S. Herbert, M. Lux, D. Wintergerst, V. Bischoff, R. Krishna, Y. Hajime, J. Groß, A. Spruss, M. Erdeljan, S. Wöll, T. Bukur, H. Muramatsu and M. Baiersdörfer for technical support, NIH Tetramer Core Facility for providing gp70 MHC class I tetramer, A. Kong for critical reading, A. Kemmer-Brück, D. Schwarck, S. Bolte for clinical operations support and K. Kariko for advice. This work was supported by the technology innovation program of the Rhineland Palatinate government, the InnoTop program, the CI3 Cutting Edge Cluster Funding of the German Ministry of Technology (BMBF) and the Collaborative Research Group 1066 of Deutsche Forschungsgemeinschaft (DFG). L.M.K. was funded by the Graduate School of Immunotherapy 1043 of DFG.
Author information
Authors and Affiliations
Contributions
U.S. was responsible for conception and experimental strategy of the study. Formulation development was performed by H.H. and P.L. Design and analysis of the experiments were done by L.M.K, M.D., H.H., S.K, M.M. and D.F. supported by K.C.R. and F.V. L.M.K, M.D., M.M., D.F., K.C.R., A.S., F.V., Y.H., Ho.He, C.G. and M.V. performed the experiments and acquired the data. C.L., J. H., J.D., A.N.K., J.B., R.J., S.H., S.G., E.D., R.R. and S.A. were involved in design, implementation or laboratory analyses of the clinical study. L.M.K., M.D., Ö.T. and U.S. interpreted the data and drafted the manuscript. C.H. supported the revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
U.S., H.H, M.M., D.F., K.C.R, C.G., Ho.He, Y.H., M.V., A.N.K., J.B., E.D., J.D., R.J. and S.H. are employees at BioNTech AG (Mainz, Germany). M.D. and S.K. are working as consultants for BioNTech AG (Mainz, Germany). U.S., L.M.K., M.D., H.H., S.K., D.F., M.M. and K.C.R. are inventors on patents and patent applications related to this study. U.S. and C.H are stock owners, C.H. is advisor and supervisory board member and U.S. is management board member of BioNTech AG. All other authors have no potential conflict of interest.
Additional information
Reviewer Information: Nature thanks O. Farokhzad, C. G. Figdor, C. Melief, L. Zhang and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
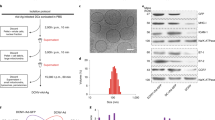
Extended Data Figure 1 Physicochemical characteristics and biological activity of RNA-LPX constituted from different lipids at various charge ratios.
a, Bioluminescence imaging of Luc expression in BALB/c mice 6 h after i.v. injection of different transfection reagents and controls: PBS (n = 3), 60 μg Luc-RNA alone (n = 3), 25 μg Luc-RNA complexed with TransMessenger (Qiagen) (n = 3), 5 μg Luc-RNA complexed with Viromer RED (Lipocalyx) (n = 3). b, Cryo-TEM images of Luc-LPX constituted at a positive:negative ((+):(−)) charge ratio of 1.3:2 with DOTMA/DOPE liposomes. Scale bar, 100 nm. c, Fraction of uncomplexed RNA in Luc-LPX preparations constituted at different charge ratios with DOTMA/DOPE liposomes determined by capillary gel electrophoresis (n = 2–7). d, Particle size, polydispersity index (left) and zeta potential (right) (n = 3) of RNA-LPX constituted with Luc-RNA and differently constituted liposomes at various charge ratios. e, Bioluminescence imaging of BALB/c mice (n = 3) after i.v. injection of Luc-LPX constituted with different liposomes at various charge ratios corresponding to d. Pie charts show relative contribution of each organ to total signal. f, Relative biodistribution of Luc expression in explanted organs of BALB/c mice (n = 3) after i.v. injection of Luc-LPX constituted with DOTMA/DOPE liposomes at a charge ratio of (+):(-) of 1.3:2 or Luc-RNA alone. g, Luc expression in human immature DCs transfected with 5 μg Luc-LPX constituted freshly or stored after constitution for indicated time periods at 4 °C (left) or room temperature (right). RNA-LPX tested in duplicates (stored) or quadruplets (fresh). Each bar represents triplicates. h, Particle size (upper left) and percentage of RNA integrity (upper right) of Luc-LPX (n = 1) incubated in 50% mouse serum for indicated time periods at 37 °C. Bioluminescence imaging of Luc expression in BALB/c mice (n = 5) after i.v. injection of Luc-LPX preincubated in 50% mouse serum for 30 min at 37 °C (lower left and right). NM, not measured. Error bars, median with interquartile range (h), otherwise mean ± s.d.
Extended Data Figure 2 Biodistribution and cellular uptake mechanism of RNA-LPX vaccines.
a, Uptake of Cy5-labelled RNA in splenic cell subsets of C57BL/6 mice (n = 3) 1 h after i.v. injection of 40 μg Cy5-labelled RNA-LPX. b, Localization of CD11c and Cy3 double-positive cells in the spleen of BALB/c mice (n = 2) 1 h after i.v. injection of 40 μg Cy3-labelled RNA-LPX. Nuclear staining in blue. Scale bar,100 μm. c, Half-life of RNA-LPX in circulation analysed by quantitative RT–qPCR in male and female C57BL/6 mice (n = 5 per time-point) after injection of 60 μg RNA-LPX constituted with NY-ESO-I, tyrosinase, MAGE-A3 and TPTE RNA (15 μg each). d, Localization of Cy5+ (upper left) or Thy1.1+ cells (lower left) in spleen and liver of BALB/c mice (n = 5) determined by microscopy or flow cytometry 1 h or 20 h after i.v. injection of 40 μg Cy5-labelled RNA-LPX or 40 μg 1-methyl-pseudouridine-modified Thy1.1-LPX, respectively. Nuclear staining in blue. Scale bar, 50 μm (top), 20 μm (bottom). Biodistribution of Cy5 signal in homogenized organs of BALB/c mice (n = 2) (right). Note the signal in the liver is overestimated in this analysis owing to the strong signal in the gall bladder, probably reflecting biliary secreted free dye. e, Bioluminescence imaging of lymph nodes of BALB/c mice (n = 3) 18 h after i.v. injection of 40 μg 1-methyl-pseudouridine-modified Luc-LPX. ax, axillary; ing, inguinal; mand, mandibular. f, Flow cytometry analysis of Cy5 and Thy1.1 expression in CD11c+ cells in the bone marrow of C57BL/6 mice (n = 3) 1 h or 20 h after i.v. injection of 40 μg Cy5-labelled RNA-LPX or 40 μg 1-methyl-pseudouridine-modified Thy1.1-LPX, respectively. g, h, Localization of Cy3-labelled RNA in human immature DCs after co-transfection of 1.25 μg Cy3-labelled RNA-LPX at a charge ratio of (+):(−) of 1.3:2 and 3:1 with dextran (g) or of 1.3:2 after staining for TLR7 or EEA1 (h). Nuclear staining in blue. Scale bar, 10 μm. i, Visualization and quantification of inhibited uptake of positively as well as negatively charged Cy3-labelled RNA-LPX in human immature DCs pretreated with rottlerin or cytochalasin D. Scale bar, 10 μm. j, Bioluminescence imaging of lymph nodes of BALB/c mice (n = 3) injected intranodally with 10 μM rottlerin in 10 μl PBS 15 min before i.v. injection of 80 μg Luc-LPX. k, Luminescence assay of whole blood enriched or not enriched with human immature DCs pretreated with poly I:C or not (control) before transfection with Luc-LPX at a charge ratio of 1.3:2. WB, whole blood. l, Poly-I:C-induced maturation determined by CD86 expression (left), bioluminescence imaging (middle) and eGFP expression in splenic cDC subsets (right) upon injection of BALB/c mice (n = 3) with 50 μg poly I:C i.p. 12 h before i.v. injection of 20 μg Luc-LPX or 80 μg eGFP-LPX, respectively. Significance was determined using unpaired two-tailed Student’s t-test (d, lower left, f, l, middle) and one-way ANOVA and Tukey’s multiple comparisons test (i–k, l, right). Error bars, mean ± s.e.m. (k) or mean ± s.d. otherwise.
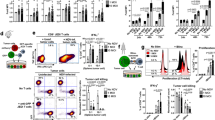
Extended Data Figure 3 Systemic TLR7- and IFNAR-dependent activation of APCs and effector cells, IFNα production and strong expansion of fully functional antigen-specific T cells induced by RNA-LPX vaccines.
a, Localization of splenic CD11chi cells at baseline (top) and 6 h after i.v. injection of 40 μg HA-LPX (bottom) into BALB/c mice (n = 2). Nuclear staining in blue. Scale bar, 100 μm. RP, red pulp; WP, white pulp. b–e, Activation marker expression in splenic cell subsets and kinetics of IFNα serum levels after i.v. injection of mice (n = 3 per time point) with HA-LPX in Tlr3−/−, Tlr4−/− and Tlr9−/− mice (b), in Ifnar1−/− mice (c, d), or in BALB/c mice treated with 100 μg anti-IFNAR1 antibody or isotype i.p. 1 h before i.v. injection of HA-LPX (e). Ab, antibody. f, mRNA levels of IFNα isoforms in sorted splenic APC subsets of C57BL/6 mice (n = 3) 1 h after i.v. injection of HA-LPX determined by qRT–PCR. Data expressed as log2-fold change, as compared to control animals. g, IFNα serum levels after i.v. injection of HA-LPX in BDCA2-DTR mice (n = 3 per time point) depleted (depl) of pDCs (left) and in C57BL/6 mice (n = 3 per time point) depleted of macrophages (right). h, CFSE proliferation profile of HA-specific CD4+ T cells in lymphoid compartments of BALB/c Thy1.1+ mice (n = 3) after adoptive transfer of HA-specific Thy1.2+ HA-TCR-transgenic CD4+ T cells and subsequent immunization with HA-LPX or control (untreated). Fraction of proliferated cells indicated. tg, transgenic. i, Priming of naive HA-specific CD8+ T cells ex vivo. BALB/c (n = 3) mice were immunized with 80 μg HA-LPX, irrelevant (eGFP)-LPX or NaCl (control). Splenocytes were prepared 12 h later and co-incubated with CFSE-labelled CL4-TCR-transgenic CD8+ T cells isolated using MACS magnetic microbeads coated with CD8 antibodies at an effector:target ratio of 1:6. Four days later, proliferation profiles were analysed by flow cytometry. Numbers indicate the percentage of proliferated cells. j, Fraction of cytokine-secreting CD8+ T cells within CD8+ T cells in the spleen upon de novo priming in C57BL/6 mice (n = 5) immunized i.v. (day 0, 3, 8) with OVA-LPX after in vitro restimulation with no (none), irrelevant VSV (irrelevant) or OVA peptide and intracellular cytokine staining (top). Spleen ex vivo ELISPOT assay upon de novo priming in BALB/c mice (n = 5) immunized i.v. (day 0, 3, 8) with gp70-LPX. Stimulation with no (none), irrelevant HA (irrelevant) or gp70 peptide (lower left). gp70-specific cytotoxicity in vivo (lower right). BALB/c mice (n = 5) were immunized i.v. (day 0, 3, 8) with 40 μg gp70-LPX. Naive splenocytes were labelled with 0.5 or 5 μM CFSE and pulsed with peptide (6 μg ml−1) five days after the last immunization, and target cells (2 × 107) were adoptively transferred into immunized recipients i.v. (irrelevant HA-loaded CFSElow:gp70-loaded CFSEhigh = 1:1). Recipient splenocytes were analysed by flow cytometry 18 h after transfer, and antigen-specific lysis was determined: specific lysis (%) = (1 − (percentage of cells pulsed with gp70/percentage of cells pulsed with HA)) × 100). k, Expression of memory markers CD127 and CD62L in gp70-specific, CD44+CD8+ T cells compared to non-specific CD8+ T cells in blood (day 19) and spleen (day 67) of BALB/c mice (n = 3) after priming with gp70-LPX (day 0, 7, 14). l, Fraction of gp70-specific CD8+ T cells within total CD8+ T cells in blood, bone marrow and lymph nodes determined by MHC class I tetramer staining after de novo priming of splenectomized BALB/c mice (n = 5–7) immunized with gp70-LPX (day 0, 7) or left untreated (control). Significance was determined using unpaired two-tailed Student’s t-test (b left, c), two-way ANOVA and Bonferroni’s multiple comparisons test (b right, g) and one-way ANOVA and Tukey’s multiple comparisons test (j, l). Error bars, mean ± s.d.
Extended Data Figure 4 Potent antitumour immunity and rejection of advanced aggressively growing tumours in mice conferred by RNA-LPX vaccines.
a, B16-OVA melanoma load in lungs of C57BL/6 mice (n = 8) immunized i.v. (days 4, 7, 11) with OVA-LPX or irrelevant (eGFP)-LPX. b, Expression of activation markers measured 24 h after i.v. injection of 40 μg irrelevant (empty vector)-LPX, eGFP-LPX or OVA-LPX by flow cytometry in splenic immune cell subsets (n = 3) and IFNα serum levels (n = 3) 6 h after injection in C57BL/6 mice. c, Bioluminescence signal of tumours in different groups before immunization and on day 25 (upper left), tumour load and lung weights (upper right) and TRP-1-specific CD8+ and CD4+ T-cell responses in spleens of control (untreated), irrelevant (empty vector)-LPX and TRP-1-LPX-immunized B6 albino mice (n = 12) on day 25 detected by ELISPOT assay (bottom), depicted in Fig. 3b. d, Bioluminescence imaging of CT26-Luc carcinoma in BALB/c mice (n = 4–7) depicted in Fig. 3c (left). e, TC-1-Luc tumour growth in C57BL/6 mice (n = 10) (left), depicted in Fig. 3d, and remission of established advanced TC-1-Luc tumours in C57BL/6 mice (n = 10) immunized i.v. with 40 μg E6/E7-LPX (day 13, 20, 27) (right). f, Survival of BALB/c mice rechallenged with CT26-Luc colon carcinoma cells on day 109, depicted in Fig. 3e. Significance was determined using one-way ANOVA and Tukey’s multiple comparisons test (c), two-way ANOVA and Bonferroni’s multiple comparisons test (d), paired two-tailed Student’s t-test (f, right), unpaired two-tailed Student’s t-test (f, far right), and log-rank test (f, left). Error bars, median with interquartile range (d), mean ± s.d. otherwise.
Extended Data Figure 5 Clinical application of RNA-LPX vaccines and de novo priming and amplification of patient T-cell responses against encoded vaccine antigens.
a, Vaccination scheme and monitoring for patients 1–3. b, Antigen-specific T-cell responses against NY-ESO-1 and tyrosinase determined by restimulation with overlapping peptide mixtures in IFNγ ELISPOT for patient 1. c, Antigen-specific T-cell responses against NY-ESO-1 and MAGE-A3, determined by post-IVS IFNγ ELISPOT assay at indicated days for patient 2. Values are corrected for background (no peptide). d, Antigen-specific T-cell responses against NY-ESO-I and MAGE-A3, determined by ex vivo IFNγ ELISPOT assay at indicated days for patient 3. Numbers in ELISPOT data indicate the amino acid position of each epitope. Significance was determined using unpaired two-tailed Student’s t-test. Error bars, mean ± s.e.m.
Extended Data Figure 6 Comparison of i.v. and s.c. routes for RNA-LPX administration in the context of T-cell priming and biodistribution of RNA-LPX upon s.c. administration.
Fraction of OVA-specific CD8+ T cells within CD8+ T cells on day 13 in blood after de novo priming of C57BL/6 mice (n = 5) immunized i.v. with OVA-LPX (day 0, 3, 8) (left). Biodistribution of Luc expression 24 h after s.c. injection of Luc-LPX in BALB/c mice (n = 3) (right). Signal can only be observed at the injection site and the draining lymph node. Significance was determined using one-way ANOVA and Tukey’s multiple comparisons test. Error bars, mean ± s.d.
Rights and permissions
About this article
Cite this article
Kranz, L., Diken, M., Haas, H. et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401 (2016). https://doi.org/10.1038/nature18300
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18300
This article is cited by
-
Nucleic acid-based drugs for patients with solid tumours
Nature Reviews Clinical Oncology (2024)
-
Dendritic cells as orchestrators of anticancer immunity and immunotherapy
Nature Reviews Clinical Oncology (2024)
-
Natural long-chain saturated fatty acids doped LNPs enabling spleen selective mRNA translation and potent cancer immunotherapy
Nano Research (2024)
-
mRNA nanodelivery systems: targeting strategies and administration routes
Biomaterials Research (2023)
-
Passive, active and endogenous organ-targeted lipid and polymer nanoparticles for delivery of genetic drugs
Nature Reviews Materials (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.