Abstract
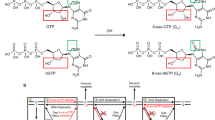
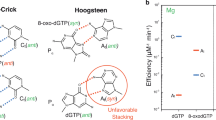
Oxidative stress promotes genomic instability and human diseases1. A common oxidized nucleoside is 8-oxo-7,8-dihydro-2′-deoxyguanosine, which is found both in DNA (8-oxo-G) and as a free nucleotide (8-oxo-dGTP)2,3. Nucleotide pools are especially vulnerable to oxidative damage4. Therefore cells encode an enzyme (MutT/MTH1) that removes free oxidized nucleotides. This cleansing function is required for cancer cell survival5,6 and to modulate Escherichia coli antibiotic sensitivity in a DNA polymerase (pol)-dependent manner7. How polymerases discriminate between damaged and non-damaged nucleotides is not well understood. This analysis is essential given the role of oxidized nucleotides in mutagenesis, cancer therapeutics, and bacterial antibiotics8. Even with cellular sanitizing activities, nucleotide pools contain enough 8-oxo-dGTP to promote mutagenesis9,10. This arises from the dual coding potential where 8-oxo-dGTP(anti) base pairs with cytosine and 8-oxo-dGTP(syn) uses its Hoogsteen edge to base pair with adenine11. Here we use time-lapse crystallography to follow 8-oxo-dGTP insertion opposite adenine or cytosine with human pol β, to reveal that insertion is accommodated in either the syn- or anti-conformation, respectively. For 8-oxo-dGTP(anti) insertion, a novel divalent metal relieves repulsive interactions between the adducted guanine base and the triphosphate of the oxidized nucleotide. With either templating base, hydrogen-bonding interactions between the bases are lost as the enzyme reopens after catalysis, leading to a cytotoxic nicked DNA repair intermediate. Combining structural snapshots with kinetic and computational analysis reveals how 8-oxo-dGTP uses charge modulation during insertion that can lead to a blocked DNA repair intermediate.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Reuter, S., Gupta, S. C., Chaturvedi, M. M. & Aggarwal, B. B. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic. Biol. Med. 49, 1603–1616 (2010)
Bont, R. D. & Larebeke, N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis 19, 169–185 (2004)
Fraga, C. G., Shigenaga, M. K., Park, J. W., Dega, P. & Ames, B. N. Oxidative damage to DNA during aging:8-hydroxy-2′-deoxyguanosine in rat organ DNA and urine. Proc. Natl Acad. Sci. USA 87, 4533–4537 (1990)
Topal, M. D. & Baker, M. S. DNA precursor pool: a significant target for N-methyl-N-nitrosourea in C3H/10T1/2 clone 8 cells. Proc. Natl Acad. Sci. USA 79, 2211–2215 (1982)
Huber, K. V. et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature 508, 222–227 (2014)
Gad, H. et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature 508, 215–221 (2014)
Foti, J. J., Devadoss, B., Winkler, J. A., Collins, J. J. & Walker, G. C. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science 336, 315–319 (2012)
Shibutani, S., Takeshita, M. & Grollman, A. P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349, 431–434 (1991)
Colussi, C. et al. The mammalian mismatch repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool. Curr. Biol. 12, 912–918 (2002)
Pursell, Z. F., McDonald, J. T., Mathews, C. K. & Kunkel, T. A. Trace amounts of 8-oxo-dGTP in mitochondrial dNTP pools reduce DNA polymerase γ replication fidelity. Nucleic Acids Res. 36, 2174–2181 (2008)
Oda, Y. et al. NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res. 19, 1407–1412 (1991)
Amouroux, R., Campalans, A., Epe, B. & Radicella, J. P. Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic Acids Res. 38, 2878–2890 (2010)
Cabelof, D. C., Raffoul, J. J., Yanamadala, S., Guo, Z. & Heydari, A. R. Induction of DNA polymerase β-dependent base excision repair in response to oxidative stress in vivo. Carcinogenesis 23, 1419–1425 (2002)
Beard, W. A. & Wilson, S. H. Structure and mechanism of DNA polymerase β. Biochemistry 53, 2768–2780 (2014)
Donigan, K. A. et al. Human POLB gene is mutated in high percentage of colorectal tumors. J. Biol. Chem. 287, 23830–23839 (2012)
Beese, L. S. & Steitz, T. A. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 10, 25–33 (1991)
Sawaya, M. R., Prasad, P., Wilson, S. H., Kraut, J. & Pelletier, H. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry 36, 11205–11215 (1997)
Batra, V. K. et al. Magnesium induced assembly of a complete DNA polymerase catalytic complex. Structure 14, 757–766 (2006)
Batra, V. K. et al. Mutagenic conformation of 8-oxo-7,8-dihydro-2′-dGTP in the confines of a DNA polymerase active site. Nature Struct. Mol. Biol. 17, 889–890 (2010)
Miller, H., Prasad, R., Wilson, S. H., Johnson, F. & Grollman, A. P. 8-OxodGTP incorporation by DNA polymerase β is modified by active-site residue Asn279. Biochemistry 39, 1029–1033 (2000)
Wang, Y. L. & Schlick, T. Distinct energetics and closing pathways for DNA polymerase β with 8-oxoG template and different incoming nucleotides. BMC Struct. Biol. 7, 7 (2007)
Freudenthal, B. D., Beard, W. A. & Wilson, S. H. DNA polymerase minor groove interactions modulate mutagenic bypass of a templating 8-oxoguanine lesion. Nucleic Acids Res. 41, 1848–1858 (2013)
Brown, J. A., Duym, W. W., Fowler, J. D. & Suo, Z. Single-turnover kinetic analysis of the mutagenic potential of 8-oxo-7,8-dihydro-2′-deoxyguanosine during gap-filling synthesis catalyzed by human DNA polymerases λ and β. J. Mol. Biol. 367, 1258–1269 (2007)
Eckenroth, B. E., Fleming, A. M., Sweasy, J. B., Burrows, C. J. & Doublie, S. Crystal structure of DNA polymerase β with DNA containing the base lesion spiroiminodihydantoin in a templating position. Biochemistry 53, 2075–2077 (2014)
Koag, M. C., Min, K. & Lee, S. Structural basis for promutagenicity of 8-halogenated guanine. J. Biol. Chem. 289, 6289–6298 (2014)
Freudenthal, B. D., Beard, W. A., Shock, D. D. & Wilson, S. H. Observing a DNA polymerase choose right from wrong. Cell 154, 157–168 (2013)
Nakamura, T., Zhao, Y., Yamagata, Y., Hua, Y.-j. & Yang, W. Watching DNA polymerase η make a phosphodiester bond. Nature 487, 196–201 (2012)
Beard, W. A. & Wilson, S. H. Structural insights into the origins of DNA polymerase fidelity. Structure 11, 489–496 (2003)
Sucato, C. A. et al. DNA polymerase β fidelity: halomethylene-modified leaving groups in pre-steady-state kinetic analysis reveal differences at the chemical transition state. Biochemistry 47, 870–879 (2008)
Harris, J. L. et al. Aprataxin, poly-ADP ribose polymerase 1 (PARP-1) and apurinic endonuclease 1 (APE1) function together to protect the genome against oxidative damage. Hum. Mol. Genet. 18, 4102–4117 (2009)
Beard, W. A. & Wilson, S. H. Purification and domain-mapping of mammalian DNA polymerase β. Methods Enzymol. 262, 98–107 (1995)
Otwinowski, Z. & Minor, W. Processsing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
The AxPyMOL Molecular Graphics Plugin for Microsoft PowerPoint. v.1.0 (Schrodinger, 2010)
Beard, W. A., Shock, D. D. & Wilson, S. H. Influence of DNA structure on DNA polymerase β active site function: extension of mutagenic DNA intermediates. J. Biol. Chem. 279, 31921–31929 (2004)
Beard, W. A., Shock, D. D., Yang, X.-P., DeLauder, S. F. & Wilson, S. H. Loss of DNA polymerase β stacking interactions with templating purines, but not pyrimidines, alters catalytic efficiency and fidelity. J. Biol. Chem. 277, 8235–8242 (2002)
Frisch, M. J. et al. Gaussian 09, Revision D.01 (Gaussian, 2009)
Chirlian, L. E. & Francl, M. M. Atomic charges derived from electrostatic potentials: a detailed study. J. Comput. Chem. 8, 894–905 (1987)
Brooks, B. R. et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983)
Feller, S. E., Zhang, Y., Pastor, R. W. & Brooks, B. R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 103, 4613–4621 (1995)
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993)
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005)
Foloppe, N. & MacKerell, J. A. D. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 21, 86–104 (2000)
MacKerell, A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998)
MacKerell, A. D. & Banavali, N. K. All-atom empirical force field for nucleic acids: II. Application to molecular dynamics simulations of DNA and RNA in solution. J. Comput. Chem. 21, 105–120 (2000)
Wang, Y., Arora, K. & Schlick, T. Subtle but variable conformational rearrangements in the replication cycle of Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) may accommodate lesion bypass. Protein Sci. 15, 135–151 (2006)
Pavelites, J. J., Gao, J., Bash, P. A. & Mackerell, A. D. A molecular mechanics force field for NAD+, NADH, and the pyrophosphate groups of nucleotides. J. Comput. Chem. 18, 221–239 (1997)
Acknowledgements
We thank the Collaborative Crystallography group at the National Institute of Environmental Health Sciences for help with data collection and analysis. We thank L. Pedersen for discussions. Use of the advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract W-31-109-Eng-38. This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (project numbers Z01-ES050158 (to S.W.), Z01-ES050161 (to S.W.), and ZIC-ES043010 (to L.P.)) and in association with National Institutes of Health grant 1U19CA105010. We are grateful for computational support for the molecular dynamics simulations from the HPC clusters at NYU as well as the Blue Gene at CCNI. Support from Philip Morris USA Inc. and Philip Morris International to T.S. is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
B.F., W.B., and S.W. designed the project. B.F. performed crystallography. D.S. did the kinetic analyses. T.K. and T.S. did the molecular dynamics simulations. L.P. did the quantum mechanical analysis. B.F., W.B., and S.W. prepared the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Mutagenic 8-oxo-dGTP insertion opposite adenine.
a, Overlay of the ternary complex for 8-oxo-dGTP(syn):Ad generated with Ca2+ or a dideoxy-terminated primer (Protein Data Bank accession number 3MBY) is shown in yellow and green, respectively (root mean squared deviation of 0.17 Å). b, The pol β active site is shown with a 2Fo − Fc map contoured at 1.5σ after a 20 s soak. Key active site residues are indicated and Mg2+ ions are shown as red spheres. The reactant 8-oxo-dGTP and product 8-oxo-dGMP are shown in green and yellow respectively. c, A focused view of b with the density removed. Coordinating waters (blue) and their distances (in ångströms) to active site metals are shown. d, The active site following a 40 s soak is shown with an omit map (3σ) for the Mgp and coordinating waters. e, The coordination distances (in ångströms) for the Nac, Mgp, and Mgn metals are indicated for the closed product complex after a 40 s soak. f, The 8-oxo-dGMP(anti):Ad contact between N3 and N6 of 8-oxo-dGMP and Ad respectively is shown for the open product complex after a 90 s soak.
Extended Data Figure 2 Pre-catalytic ground state with 8-oxo-dGTP and templating cytosine after a 5 s soak in MnCl2.
a, The pre-catalytic pol β active site is shown with an omit map (3σ). The ground state metal (Mng) has been removed for clarity. b, The view is a 90° rotation relative to a. An overlay of the 8-oxo-dGTP(anti) with Ca2+ and Mn2+ is shown in yellow and purple respectively. The anomalous density map contoured at 5σ for the Mn2+ ions is shown in purple. The Mng coordinating water molecules are shown in blue and the distances (in ångströms) are indicated.
Extended Data Figure 3 Reaction with 8-oxo-dGTP opposite templating cytosine.
a, Focused view of the active site following a 40 s soak is shown with key residues indicated; density has been removed for clarity (see Fig. 2e for density). b, An omit map (3σ) for Cag is shown. Coordinating waters are shown in blue (distances in ångströms). c, An omit map (3σ) for Mgp is shown.
Extended Data Figure 4 Quantum mechanical computational models for 8-oxo-dGTP(anti) and dGTP(anti).
The models used for the quantum mechanical computational studies with the calcium ions, oxygen, phosphates, carbon, nitrogen, and protons shown in green, red, orange, grey, blue, and white, respectively. The key atoms and Asp 190, Asp 192, and Asp 256 mimics are indicated. a, The 8-oxo-dGTP(anti) with three calcium ions and eight water molecules. b, The 8-oxo-dGTP(anti) with two calcium ions and three water molecules. c, The dGTP(anti) with two calcium ions and three water molecules.
Extended Data Figure 5 Molecular dynamics simulation analysis of 8-oxo-dGTP(anti) and dGTP(anti) opposite Cy.
a, The 8-oxo-dGTP(anti) opposite Cy at 80 ns superimposed upon the initial structure. A multicolour code based on atom type is used for the final molecular dynamics structure, whereas the reference initial structure is shown in light grey. The catalytic (Mgc), nucleotide (Mgn), and ground (Mgg) magnesium metal ions are shown in green, and average distances over the course of the simulation are indicated for Pα-O3′ and Mgc-O3′. b, Distance distributions between hydrogen atoms in the water shell and O8 in the 8-oxo-dGTP(anti):Cy simulation. A snapshot of the 8-oxo-dGTP, Mgg, and water shell (W(g1–g5)) is plotted at top. Black and red dotted lines indicate Mgg coordination and a hydrogen-bonding interaction between a water molecule and O8, respectively. Four of the five water molecules in the water shell (W(g1–g4)) contribute to hydrogen-bonding interactions with O8. Blue and orange lines indicate distances between hydrogen atoms in each water molecule and O8. The red line in the bottom plot indicates the minimum distance between hydrogen atoms in the water shell and O8. c, The dGTP(anti) opposite Cy at 80 ns superimposed upon the initial structure (grey). Distances and ion labelling are as for a. d, Root mean squared deviation of the evolving molecular dynamics structure for the entire polymerase/DNA complex (top) and for the active site only (bottom), with respect to the crystal structure.
Rights and permissions
About this article
Cite this article
Freudenthal, B., Beard, W., Perera, L. et al. Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide. Nature 517, 635–639 (2015). https://doi.org/10.1038/nature13886
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13886
This article is cited by
-
Widespread 8-oxoguanine modifications of miRNA seeds differentially regulate redox-dependent cancer development
Nature Cell Biology (2023)
-
In crystallo observation of three metal ion promoted DNA polymerase misincorporation
Nature Communications (2022)
-
8-Oxoguanine: from oxidative damage to epigenetic and epitranscriptional modification
Experimental & Molecular Medicine (2022)
-
Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints
Nature Communications (2022)
-
Biology of aging: Oxidative stress and RNA oxidation
Molecular Biology Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.