Abstract
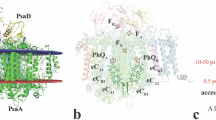
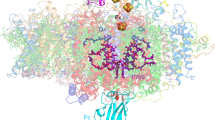
Photosynthesis converts light energy into biologically useful chemical energy vital to life on Earth. The initial reaction of photosynthesis takes place in photosystem II (PSII), a 700-kilodalton homodimeric membrane protein complex that catalyses photo-oxidation of water into dioxygen through an S-state cycle of the oxygen evolving complex (OEC). The structure of PSII has been solved by X-ray diffraction (XRD) at 1.9 ångström resolution, which revealed that the OEC is a Mn4CaO5-cluster coordinated by a well defined protein environment1. However, extended X-ray absorption fine structure (EXAFS) studies showed that the manganese cations in the OEC are easily reduced by X-ray irradiation2, and slight differences were found in the Mn–Mn distances determined by XRD1, EXAFS3,4,5,6,7 and theoretical studies8,9,10,11,12,13,14. Here we report a ‘radiation-damage-free’ structure of PSII from Thermosynechococcus vulcanus in the S1 state at a resolution of 1.95 ångströms using femtosecond X-ray pulses of the SPring-8 ångström compact free-electron laser (SACLA) and hundreds of large, highly isomorphous PSII crystals. Compared with the structure from XRD, the OEC in the X-ray free electron laser structure has Mn–Mn distances that are shorter by 0.1–0.2 ångströms. The valences of each manganese atom were tentatively assigned as Mn1D(iii), Mn2C(iv), Mn3B(iv) and Mn4A(iii), based on the average Mn–ligand distances and analysis of the Jahn–Teller axis on Mn(iii). One of the oxo-bridged oxygens, O5, has significantly longer distances to Mn than do the other oxo-oxygen atoms, suggesting that O5 is a hydroxide ion instead of a normal oxygen dianion and therefore may serve as one of the substrate oxygen atoms. These findings provide a structural basis for the mechanism of oxygen evolution, and we expect that this structure will provide a blueprint for the design of artificial catalysts for water oxidation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Umena, Y., Kawakami, K., Shen, J.-R. & Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 (2011)
Yano, J. et al. X-ray damage to the Mn4Ca complex in single crystals of photosystem II: a case study for metalloprotein crystallography. Proc. Natl Acad. Sci. USA 102, 12047–12052 (2005)
Yano, J. et al. High-resolution Mn EXAFS of the oxygen-evolving complex in photosystem II: structural implications for the Mn4Ca cluster. J. Am. Chem. Soc. 127, 14974–14975 (2005)
Yano, J. et al. Where water is oxidized to dioxygen: structure of the photosynthetic Mn4Ca cluster. Science 314, 821–825 (2006)
Dau, H. & Haumann, M. The manganese complex of photosystem II in its reaction cycle — basic framework and possible realization at the atomic level. Coord. Chem. Rev. 252, 273–295 (2008)
Dau, H., Grundmeier, A., Loja, P. & Haumann, M. On the structure of the manganese complex of photosystem II: extended-range EXAFS data and specific atomic-resolution models for four S-states. Phil. Trans. R. Soc. Lond. B 363, 1237–1243 (2008)
Glöckner, C. et al. Structural changes of the oxygen-evolving complex in photosystem II during the catalytic cycle. J. Biol. Chem. 288, 22607–22620 (2013)
Ames, W. et al. Theoretical evaluation of structural models of the S2 state in the oxygen evolving complex of Photosystem II: protonation states and magnetic interactions. J. Am. Chem. Soc. 133, 19743–19757 (2011)
Galstyan, A., Robertazzi, A. & Knapp, E. W. Oxygen-evolving Mn cluster in photosystem II: the protonation pattern and oxidation state in the high-resolution crystal structure. J. Am. Chem. Soc. 134, 7442–7449 (2012)
Grundmeier, A. & Dau, H. Structural models of the manganese complex of photosystem II and mechanistic implications. Biochim. Biophys. Acta 1817, 88–105 (2012)
Isobe, H. et al. Theoretical illumination of water-inserted structures of the CaMn4O5 cluster in the S2 and S3 states of oxygen-evolving complex of photosystem II: full geometry optimizations by B3LYP hybrid density functional. Dalton Trans. 41, 13727–13740 (2012)
Luber, S. et al. S1-state model of the O2-evolving complex of photosystem II. Biochemistry 50, 6308–6311 (2011)
Pantazis, D. A., Ames, W., Cox, N., Lubitz, W. & Neese, F. Two interconvertible structures that explain the spectroscopic properties of the oxygen-evolving complex of photosystem II in the S2 state. Angew. Chem. Int. Edn Engl. 51, 9935–9940 (2012)
Blomberg, M. R. A., Borowski, T., Himo, F., Liao, R. Z. & Siegbahn, P. E. M. Quantum chemical studies of mechanisms for metalloenzymes. Chem. Rev. 114, 3601–3658 (2014)
Zouni, A. et al. Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature 409, 739–743 (2001)
Kamiya, N. & Shen, J.-R. Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-Å resolution. Proc. Natl Acad. Sci. USA 100, 98–103 (2003)
Ferreira, K. N., Iverson, T. M., Maghlaoui, K., Barber, J. & Iwata, S. Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838 (2004)
Guskov, A. et al. Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nature Struct. Mol. Biol. 16, 334–342 (2009)
Southworth-Davies, R. J., Medina, M. A., Carmichael, I. & Garman, E. F. Observation of decreased radiation damage at higher dose rates in room temperature protein crystallography. Structure 15, 1531–1541 (2007)
Neutze, R., Wouts, R., van der Spoel, D., Weckert, E. & Hajdu, J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 406, 752–757 (2000)
Kern, J. et al. Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature. Science 340, 491–495 (2013)
Kupitz, C. et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature 513, 261–265 (2014)
Kern, J. et al. Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy. Nature Commun. 5, 4371 (2014)
Holton, J. M. & Frankel, K. A. The minimum crystal size needed for a complete diffraction data set. Acta Crystallogr. D 66, 393–408 (2010)
Hirata, K. et al. Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL. Nature Methods 11, 734–736 (2014)
Shoji, M. et al. Theoretical modeling of biomolecular systems I. Large scale QM/MM calculations of hydrogen bonding networks of oxygen evolving complex of photosystem II. Mol. Phys. http://dx.doi.org/10.1080/00268976.2014.960021 (published online, 29 September 2014)
Cox, N. et al. Effect of Ca2+/Sr2+ substitution on the electronic structure of the oxygen-evolving complex of photosystem II: a combined multifrequency EPR, 55Mn-ENDOR, and DFT study of the S2 state. J. Am. Chem. Soc. 133, 3635–3648 (2011)
Rapatskiy, L. et al. Detection of the water-binding sites of the oxygen-evolving complex of Photosystem II using W-band 17O electron-electron double resonance-detected NMR spectroscopy. J. Am. Chem. Soc. 134, 16619–16634 (2012)
Siegbahn, P. E. Water oxidation mechanism in photosystem II, including oxidations, proton release pathways, O-O bond formation and O2 release. Biochim. Biophys. Acta 1827, 1003–1019 (2013)
Cox, N. et al. Electronic structure of the oxygen-evolving complex in photosystem II prior to O-O bond formation. Science 345, 804–808 (2014)
Shen, J.-R. & Kamiya, N. Crystallization and the crystal properties of the oxygen-evolving photosystem II from Synechococcus vulcanus . Biochemistry 39, 14739–14744 (2000)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Murray, J., Garman, E. & Ravelli, R. X-ray absorption by macromolecular crystals: the effect of wavelength and crystal composition on absorbed dose. J. Appl. Crystallogr. 37, 513–522 (2004)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Acknowledgements
We thank T. Ishikawa, M. Yabashi, K. Tono and Y. Inubushi for help in using the SACLA beamline, T. Tsukihara and S. Yoshikawa for suggestions and comments on the experiments, F. Seno for assistance with sample preparation, and K. Kawakami and Y. Umena for suggestions in the initial stage of the project. F. H. M. Koua participated in the initial stage of this work. This work was supported by an X-ray Free Electron Laser Priority Strategy Program (The Ministry of Education, Culture, Sports, Science and Technology of Japan, MEXT) (H.A. and J.-R.S.), a JST/CREST grant (K.H.), a grant-in-aid for Specially Promoted Research no. 24000018 (J.-R.S.) and KAKENHI grant no. 26840023 (M.S.) from JSPS, MEXT, Japan. The XFEL experiments were performed at beamline 3 of SACLA with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (proposal nos 2012A8011, 2012B8040, 2013A8047, 2013B8052 and 2014A8036), and we thank staff at SACLA for their help.
Author information
Authors and Affiliations
Contributions
H.A. and J.-R.S. planned and organized the experiments, F.A. and Y.N. prepared the samples, F.A. made the crystals, G.U., H.M., K.H., H.A. and M.Y. designed and established the experimental set-up, M.S., F.A., T.S., Y.N., G.U., H.M., K.H., H.A., M.Y. and J.-R.S. conducted the diffraction experiments, K.Y. performed scaling of raw diffraction images, M.S. performed the structural analysis, M.S. and J.-R.S. wrote the manuscript, and all authors discussed and commented on the results and the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Diffraction experiment using the XFEL beam at SACLA.
a, Schematic drawing of the diffraction experiment. Still diffraction images were recorded from different points on large PSII crystals. The crystals were rotated by 0.2° between each image over a range of 180°. Adjacent irradiation points were separated by 50 µm in the horizontal direction for any rotational angle. Translation in the vertical direction was varied depending on the rotational angle, so that the irradiation points were also separated by 50 µm in the vertical direction. b, A picture of the PSII crystal in a cryo-loop after the XFEL diffraction experiment. Note that the path where the XFEL beam passed through became hollowed out, and resulted in footprints of the irradiation points, which were well separated.
Extended Data Figure 2 A diffraction image from a PSII crystal obtained with the XFEL beam.
A typical diffraction image is shown; the boxed area at the right is shown enlarged in the inset, where diffraction spots at the maximum resolution are visible.
Extended Data Figure 3 Anomalous signals of the metal ions obtained with the XFEL beam.
Shown are anomalous difference Fourier maps contoured at 5σ distributions. a, Manganese atoms of the OEC. b, The non-haem iron in the acceptor side between QA and QB.
Extended Data Figure 4 Coordination environment of Mn1D, Mn2C, Mn3B and Mn4A.
The ligand environment of a, Mn1D, b, Mn2C, c, Mn3B and d, Mn4A are drawn in stereo view. The ligand bonds involving O5 coordination are slightly longer than the others in both Mn1D and Mn4A. Note that Mn1D is in a pseudo-five-coordinated, trigonal bipyramidal geometry with additional weak interaction to O5. Colour code: grey, manganese; green, carbon; blue, nitrogen; red, oxo-oxygen; yellow, O5; orange, water.
Extended Data Figure 5 Jahn-Teller axes on Mn1D and Mn4A.
The ligand environments of Mn1D and Mn4A are drawn in a stereo view. Based on the longer ligand bonds involving the O5 coordination as shown in Extended Data Fig. 4a, d, two possible Jahn–Teller axes were assigned which are approximately parallel. Colour codes are the same as those in Extended Data Fig. 4. Light blue lines indicate two Jahn–Teller axes found in the OEC in the S1 state. a, View direction almost orthogonally oriented relative to the two Jahn–Teller axes. b, View direction rotated by 90° from a.
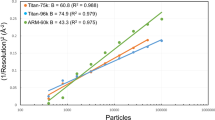
Extended Data Figure 6 Scaling of the raw images.
Standard deviations of the average pixel values calculated in the group belonging to the same crystal before and after scaling are plotted. X and Y axes indicate standard deviations obtained before and after scaling, respectively. Red line in the plot represents the relationship Y = X. a, Plot of the images collected during the first half of the beam time (including the first half of the images for data set 1). b, Plot of the images collected during the last half of the beam time (including the rest of the images for data set 1 and all of the images for data set 2).
Rights and permissions
About this article
Cite this article
Suga, M., Akita, F., Hirata, K. et al. Native structure of photosystem II at 1.95 Å resolution viewed by femtosecond X-ray pulses. Nature 517, 99–103 (2015). https://doi.org/10.1038/nature13991
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13991
This article is cited by
-
Oxygen-evolving photosystem II structures during S1–S2–S3 transitions
Nature (2024)
-
Evolutionary diversity of proton and water channels on the oxidizing side of photosystem II and their relevance to function
Photosynthesis Research (2023)
-
Absorption changes in Photosystem II in the Soret band region upon the formation of the chlorophyll cation radical [PD1PD2]+
Photosynthesis Research (2023)
-
Theoretical elucidation of the structure, bonding, and reactivity of the CaMn4Ox clusters in the whole Kok cycle for water oxidation embedded in the oxygen evolving center of photosystem II. New molecular and quantum insights into the mechanism of the O–O bond formation
Photosynthesis Research (2023)
-
Absolute quantification of cellular levels of photosynthesis-related proteins in Synechocystis sp. PCC 6803
Photosynthesis Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.