Abstract
Coccolithophores have influenced the global climate for over 200 million years1. These marine phytoplankton can account for 20 per cent of total carbon fixation in some systems2. They form blooms that can occupy hundreds of thousands of square kilometres and are distinguished by their elegantly sculpted calcium carbonate exoskeletons (coccoliths), rendering them visible from space3. Although coccolithophores export carbon in the form of organic matter and calcite to the sea floor, they also release CO2 in the calcification process. Hence, they have a complex influence on the carbon cycle, driving either CO2 production or uptake, sequestration and export to the deep ocean4. Here we report the first haptophyte reference genome, from the coccolithophore Emiliania huxleyi strain CCMP1516, and sequences from 13 additional isolates. Our analyses reveal a pan genome (core genes plus genes distributed variably between strains) probably supported by an atypical complement of repetitive sequence in the genome. Comparisons across strains demonstrate that E. huxleyi, which has long been considered a single species, harbours extensive genome variability reflected in different metabolic repertoires. Genome variability within this species complex seems to underpin its capacity both to thrive in habitats ranging from the equator to the subarctic and to form large-scale episodic blooms under a wide variety of environmental conditions.
Similar content being viewed by others
Main
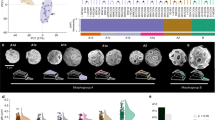
Fundamental uncertainties exist regarding the physiology and ecology of E. huxleyi, and the relationships between different morphotypes (Fig. 1a). To investigate its gene repertoire and physiological capacity, we sequenced the diploid genome of CCMP1516 using the Sanger shotgun approach. The haploid genome is estimated to be 141.7 megabases (Mb) and 97% complete on the basis of conserved eukaryotic single-copy genes5,6 (Supplementary Table 1, Supplementary Data 7 and Supplementary Information 1.1–1.4). It is dominated by repetitive elements, constituting >64% of the sequence, much greater than seen for sequenced diatoms (Fig. 2 and Supplementary Information 2.10). Of the 30,569 protein-coding genes predicted—93% of which have transcriptomic support (expressed sequence tag or RNA-seq) (Supplementary Information 1.5–1.7, 2.1–2.2 and Supplementary Data 1–3)—we identified expansions in gene families specific to iron/macromolecular transport, post-translational modification, cytoskeletal development and signal transduction relative to other sequenced eukaryotic algae (Supplementary Information 2.3).
a, E. huxleyi has five well-characterized calcification morphotypes and an overcalcified state1. b, Cladogram showing the distinct branch occupied by the haptophyte lineage on the basis of RAxML analysis of concatenated, nuclear-encoded proteins after addition of homologues from CCMP1516 and a pico-prymnesiophyte-targeted metagenome8. Lineages with algal taxa are indicated (symbol). Filled circles represent nodes with ≥70% bootstrap support. The tree is rooted for display purposes only.
Structural composition of genomes from CCMP1516 and the diatom P. tricornutum. Grey-shaded regions of each class depict proportions of tandem repeats and low-complexity regions. The grey vertical box contains only tandem repeats and low-complexity sequence. Pie charts indicate the proportion of non-repeated (white) and repeated or low-complexity (black) sequences in each haploid genome.
The E. huxleyi genome provides a crucial reference point for evolutionary, cellular and physiological studies because haptophytes represent a distinct branch on the eukaryotic tree of life (Fig. 1b). Consistent with other published analyses7, conserved marker genes demonstrate the haptophytes branch as a sister clade to heterokonts, alveolates and rhizarians. However, as a lineage possessing secondary plastids, the evolutionary history of haptophyte genomes may be more complex8 than that suggested by a single concatenated analysis. Thus, individual gene phylogenies were constructed using clusters of orthologous proteins (1,563) identified by comparative analysis of E. huxleyi and at least 9 of 48 taxa sampled from across eukaryotes (Supplementary Information 2.4). E. huxleyi was monophyletic, with heterokonts in 28–33% of the resolved trees and the green lineage (green algae and plants) in 11–14%. Less frequent relationships were also observed, presumably reflecting a mosaic genome8 with contributions from the host lineage, the eukaryotic endosymbiont, and possibly horizontal gene transfer (Supplementary Fig. 1 and Supplementary Data 4).
Coccolithophores produce the anti-stress osmolyte dimethylsulphoniopropionate (DMSP), which can be demethylated to produce methylmercaptopropionate and/or cleaved by some organisms, such as E. huxleyi, to produce the predominant natural source of atmospheric sulphur, dimethylsulphide. Although the gene encoding the DmdA protein, which catalyses the initial demethylation of DMSP, was not detected in the genome, genes that produce sulphur and carbon intermediates and function in later stages of DMSP degradation were identified9. Also present is an intron-containing, but otherwise bacterial dddD-like, gene encoding an acetyl-coenzyme A (acetyl-CoA) transferase proposed to add CoA to DMSP before cleavage9 (Supplementary Table 2). These data will facilitate molecular approaches for probing DMSP biogeochemistry and the environmental importance of sulphur production and biotransformations.
E. huxleyi synthesizes unusual lipids that are used as nutritional/feedstock supplements, polymer precursors and petrochemical replacements. Two functionally redundant pathways for the synthesis of omega-3 polyunsaturated eicosapentaenoic and docosahexaenoic fatty acids were partially characterized10 (Supplementary Table 3). Pathway analysis indicates that E. huxleyi sphingolipids are primarily glucosylceramides, often with an unusual C9 methyl branch (Supplementary Table 3) found only in fungi and some animals11. Genes for two zinc-containing quinone reductases, involved in reduction of alkenone α,β-double bonds used in paleotemperature reconstructions and proposed biofuels, were also identified12,13.
Coccoliths have precise nanoscale architecture and unique light-scattering properties of interest to material and optoelectronic scientists. Carbonic anhydrase is associated with biomineralization in other organisms14 and accelerates bicarbonate formation. The 15 E. huxleyi carbonic anhydrase isozymes and genes involved in calcium and carbon transport, H+ efflux, cytoskeleton organization and polysaccharide modulation (Supplementary Table 4) represent targets for resolving molecular mechanisms governing coccolith formation, and will aid in predicting response patterns to anthropogenic CO2 increases and ocean acidification.
The global distribution of E. huxleyi (for example, Fig. 3a, c) and its capacity for bloom formation under different physiochemical parameters are puzzling. To investigate the potential influence of genome variation in this ecological dynamic, three E. huxleyi isolates (92A, EH2 and Van556) from different oceanic regions were deeply sequenced (265–352-fold coverage) (Fig. 3a, c, Supplementary Tables 5–7 and Supplementary Information 2.6). Two approaches were used to compare genomes. First, sequence reads were assembled and contigs aligned to the CCMP1516 reference genome using Standard Nucleotide BLAST (BLASTn; Supplementary Information 2.6.1). Although these isolates show >98% 18S ribosomal RNA (rRNA) identity, only 54–77% of their contigs showed similarity to CCMP1516. 71 Mb of the remaining contigs were shared between at least two deeply sequenced strains. 8–40 Mb appeared to be isolate specific, as did 27 Mb of CCMP1516. Flow cytometric genome-size estimates also showed heterogeneity across isolates, with haploid genome sizes ranging from 99 to 133 Mb (Supplementary Information 2.5, 2.6.1 and Supplementary Table 5). These findings indicated considerable intraspecific variation.
a, Isolation locations shown over the averaged Reynolds monthly sea-surface temperature (SST) climatology (1985–2007). b, tBLASTn homology search results using predicted CCMP1516 proteins against assemblies from other strains. Bars are coloured according to the number of gene products and nucleotide per cent identity. c, Best Bayesian topology, where node values indicate posterior probability/maximum-likelihood bootstrap support. Haploid genome sizes (in Mb) are provided in brackets (with ND indicating not determined), and shaded boxes denote robust clades of geographically dispersed strains. The variable distribution of nitrite reductase (NirS) and plastocyanin (PetE) is shown.
To examine potential variations in gene content further, sequence reads were directly mapped to the CCMP1516 genome. Of the 30,569 predicted genes in CCMP1516, between 1,373 and 2,012 different genes were not found in 92A, Van556 and EH2 (cumulatively 5,218, or 17% of CCMP1516 genes), and 364 appeared to be missing from all three. These findings cannot be explained by poor coverage or sequencing bias alone. Of 458 highly conserved eukaryotic genes from the CEGMA set5, 95–97% were identified in the isolates, indicating nearly complete genome sequences (Supplementary Data 7). Together, de novo assemblies and direct mapping to CCMP1516 indicate that the pan genome of E. huxleyi represents a rapidly changing repository of genetic information with genomic fluidity estimated to be ≥10%15 (on the basis of CCMP1516 gene content).
E. huxleyi isolate differences were assessed further by Illumina sequencing of ten additional strains. Although sequenced at lower coverage, these strains were estimated to be 91–95% complete (Supplementary Tables 6, 7 and Supplementary Data 7). Direct mapping of reads from the 13 strains to CCMP1516 revealed a ‘core genome’ containing about two-thirds of the genes predicted in the reference genome (Supplementary Information 2.6.2 and Supplementary Data 5), a core independently confirmed by comparative DNA microarrays (Supplementary Information 2.7, Supplementary Data 6 and Supplementary Fig. 2). Nearly 25% of CCMP1516 genes were not found in at least three other strains, indicating that E. huxleyi represents a species complex with a genetic repertoire much greater than that of any one strain (Supplementary Figs 3, 4). Although the most extensive gene-sequence divergence was observed between CCMP1516 and deeply sequenced isolates Van556, 92A and EH2, concatenated phylogenies define three well-supported clades that are not necessarily reflective of geographic distributions (Fig. 3b, c and Supplementary Information 2.61, 2.8).
We searched the CCMP1516 genome for evidence of molecular mechanisms contributing to genome plasticity. There was limited evidence for horizontal gene transfers (Supplementary Information 2.9 and Supplementary Table 8), and although diverse, the complement of transposable elements was also small (Fig. 2 and Supplementary Information 2.10.2). However, E. huxleyi has a high density of unclassified repeats (∼31%) and tandem repeats/low-complexity regions (∼34%) with tandem-repeat/low-complexity density highest in introns (Fig. 2, Supplementary Information 2.10.1 and Supplementary Table 9). Most protein-coding genes contain multiple introns, often with noncanonical GC donor sites (Supplementary Fig. 5). The preference for 10–11-base-pair repeats in introns and their strong strandedness (meaning that on the sense and antisense strand either the motif or its reverse complement is highly favoured) raises the possibility that intronic tandem repeats have a functional role in exon swapping (Supplementary Information 2.10.3–2.10.5 and Supplementary Table 9).
E. huxleyi blooms under many different oceanographic regimes. We explored how the core genome and variable components in different ecotypes might influence success (Supplementary Information 2.11 and Supplementary Fig. 6). The remarkable capacity of E. huxleyi to withstand photoinhibition16 lies in the core genome, which encodes a variety of photoreceptors; proteins that function in the assembly and repair of photosystem II, such as D1-specific proteases and FtsH enzymes; and proteins that have a role in non-photochemical quenching (NPQ) or synthesis of NPQ compounds (Supplementary Table 10). Genes encoding reactive oxygen species (ROS) scavenging antioxidants, enzymes for synthesis of vitamin B6 constituents used during photo-oxidative stress in plants17 (Supplementary Tables 10, 15) and many light-harvesting complex (LHC) proteins are also in the core. Of the 68 LHCs, 17 belong to LI818 or LHCZ classes with photoprotective capabilities18 (Supplementary Table 11 and Supplementary Information 3.1). The complex repertoire of photoprotectors facilitates tolerance to high light by minimizing ROS accumulation and preventing oxidative damage.
Phosphorus and nitrogen are key determinants of oceanic primary production. A suite of core genes allows E. huxleyi to thrive in low phosphorus conditions. This includes six inorganic phosphate transporters (Fig. 4), a high-efficiency alkaline phosphatase (Fig. 4)19, purple acid phosphatases and other enzymes used to hydrolyse and acquire organic phosphorus compounds20. Genes for the synthesis of betaine and sulpholipids used as replacements for cellular phospholipids21 are also present (Supplementary Table 12). Numbers of phosphate transporters and alkaline phosphatases, (Fig. 4) however, vary considerably from strain to strain, supporting previous observations of differences in phosphorus uptake and hydrolysis kinetics22.
a, Key genes (gene numbers on axes) involved in nutrient acquisition and metabolism, including ammonium transporters (AMT), urea transporters (UT), nitrilase (NIT), phosphate transporters (PTA), alkaline phosphatase (PHOA), ferredoxin (FDX), flavodoxin (FldA) and nitrate reductase (NAR) (Supplementary Information 3.2). b, Genes encoding calcium EF hand (CaEF) proteins and others that bind metals such as copper, zinc and iron (Supplementary Information 3.2).
Genes for inorganic nitrogen uptake and assimilation (nitrate, nitrite and ammonium) and for acquisition and degradation of nitrogen-rich compounds (for example, urea) (Fig. 4 and Supplementary Table 13) are present in the core genome and may explain the broad range of nitrogen concentrations in which E. huxleyi blooms23. Although present in multiple copies, the number of genes encoding nitrite (4), nitrate (8) and urea (3) transporters was relatively small compared to ammonium transporters (20). This enrichment, and the varied distribution across strains (Fig. 4), may be indicative of strain-specific ammonium preference, or the need for tightly regulated transporters to mediate high-affinity ammonium/ammonia uptake while offering ammonium-toxicity protection. Surprisingly, core iron-containing (nirK) versus clade-restricted copper-containing (nirS) nitrite reductases were identified (Fig. 3), although iron is often more limiting than copper in oceanic environments.
E. huxleyi grows well in surface waters where iron levels are generally low (0.02–1 nM)24. The core genome indicates that iron is acquired using the natural resistance-associated macrophage protein (NRAMP) class of metal transporters, multicopper oxidases, surface-bound ferric reductases, and possibly, membrane-bound siderophores (Supplementary Data 8). Genes involved in mechanisms limiting iron requirements are also in the core, including manganese and copper/zinc superoxide dismutases, both zinc and iron alcohol dehydrogenases and rubredoxins, and copper- and haem- plastocyanins (PetE) and ascorbate oxidases. Selective recruitment of these enzymes as well as flavodoxin, a functional analogue of ferredoxin, may reduce iron demands25. E. huxleyi encodes many iron-binding proteins, 80 in the core and 30 linked to the variable genome (Fig. 4). Iron limitation is linked to reduced calcification and photosynthesis26, and our analysis suggests cellular demands and mechanisms to alleviate iron deprivation differ between strains and are probably important factors shaping E. huxleyi ecological dynamics.
The E. huxleyi pan genome encodes nearly 700 proteins whose structure and function is dependent upon metal binding (Supplementary Data 8). Selenium is essential for growth27 and potentially incorporated into at least 49 proteins (20 gene families) present in nearly all strains (Supplementary Table 14). Zinc affects growth and nitrogen usage26, and is a cofactor of more than 400 proteins, many present in the variable genome (Fig. 4). Heterogeneity in zinc-binding proteins across strains may explain variations in zinc quotas between cultured isolates26,28.
In addition to metals, E. huxleyi relies on a range of vitamins. Genes for de novo synthesis of antioxidants such as pro-vitamin A, vitamins C, E, B6 and B9 and the ultraviolet-light-absorbing vitamin D are uniformly present across strains. E. huxleyi, however, is ostensibly unable to inhabit ocean regions where vitamins B1 and B12 are inaccessible. ThiC, a key B1 biosynthesis enzyme, was not found in the genome, and despite relying exclusively on a vitamin-B12-dependent methionine synthase, genes for a B12 transporter and several enzymes required for B12 synthesis are also absent (Supplementary Table 15).
E. huxleyi is the dominant bloom-forming coccolithophore and can be abundant in oligotrophic oceans, directly influencing global carbon cycling. Distributions in modern oceans and those dating back to the Pleistocene era demonstrate its tremendous capacity for adaptation. Until now, the underlying mechanisms for the physiological and morphological variations between isolates have been elusive. Evidence presented here indicates that this capacity can be explained, in part, by its pan genome, the first of its kind reported for what was thought to be a single microbial eukaryotic algal species. Variations in gene complements (Fig. 4) within this species complex may drive phenotypic variation, ecological dynamics and the physiological heterogeneity observed in past studies. The high level of diversity indicates that a single strain is unlikely to be typical—or representative—of all strains. Future sequencing of phytoplankton isolates will reveal whether this discovery is a unique or more common feature in microalgae. Together, the physiological capacity and genomic plasticity of E. huxleyi make it a powerful model for the study of speciation and adaptations to global climate change.
Methods Summary
The diploid genome of CCMP1516 (isolated from the Equatorial Pacific (02.6667S 82.7167W)) was Sanger sequenced and assembled using the Arachne assembler. Gene models were predicted and validated using computational tools, experimental data (including transcriptomics; Sanger and Illumina sequenced) and NimbleGen tiling array experiments. Thirteen additional strains were sequenced using Illumina and mapped to the reference genome. A detailed description of materials and methods is in Supplementary Information.
Accession codes
Primary accessions
GenBank/EMBL/DDBJ
Referenced accessions
Sequence Read Archive
Data deposits
This paper is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence, and the online version of this paper is freely available to all readers. Assembly and annotation data for E. huxleyi strain 1516 are available through JGI Genome Portal at http://jgi.doe.gov/Ehux and at DDBJ/EMBL/GenBank under accession number AHAL00000000. The version described in this paper is the first version, AHAL01000000. Sequence information for other strains can be found at the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under the accession number SRA048733.2.
Change history
10 July 2013
Spelling of author J.N. was corrected.
References
Paasche, E. A review of the coccolithophorid Emiliania huxleyi (Prymnesiophyceae), with particular reference to growth, coccolith formation, and calcification-photosynthesis interactions. Phycologia 40, 503–529 (2001)
Poulton, A. J., Adey, T. R., Balch, W. M. & Holligan, P. M. Relating coccolithophore calcification rates to phytoplankton community dynamics: regional differences and implications for carbon export. Deep-Sea Res. II 54, 538–557 (2007)
Holligan, P. M., Viollier, M., Harbour, D. S., Campus, P. & Champagne-Philippe, M. Satellite and ship studies of coccolithophore production along a continental shelf edge. Nature 304, 339–342 (1983)
Rost, B. & Riebesell, U. in Coccolithophores: From Molecular Processes to Global Impact (eds Thierstein, H. R. & Young, J. R. ) 99–125 (Springer, 2004)
Parra, G., Bradnam, K., Ning, Z., Keane, T. & Korf, I. Assessing the gene space in draft genomes. Nucleic Acids Res. 37, 289–297 (2009)
Colbourne, J. K. et al. The ecoresponsive genome of Daphnia pulex . Science 331, 555–561 (2011)
Burki, F., Okamoto, N., Pombert, J. F. & Keeling, P. J. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc. R. Soc. B 279, 2246–2254 (2012)
Cuvelier, M. L. et al. Targeted metagenomics and ecology of globally important uncultured eukaryotic phytoplankton. Proc. Natl Acad. Sci. USA 107, 14679–14684 (2010)
Todd, J. D. et al. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315, 666–669 (2007)
Sayanova, O. et al. Identification and functional characterisation of genes encoding the omega-3 polyunsaturated biosynthetic pathway from the coccolithophore Emiliania huxleyi . Phytochemistry 72, 594–600 (2011)
Oura, T. & Kajiwara, S. Candida albicans sphingolipid C9-methyltransferase in involved in hyphal elongation. Microbiology 156, 1234–1243 (2010)
Conte, M. N., Eglinton, G. & Madureira, L. A. S. Long-chain alkenones and alkyl alkenoates as palaeotemperature indicators: their production, flux, and early sedimentary diagenesis in the Eastern North Atlantic. Advances in Organic Chemistry 19, 287–298 (1992)
Wu, Q., Shiraiwa, Y., Takeda, H., Sheng, G. & Fu, J. Liquid-saturated hydrocarbons resulting from pyrolysis of the marine Coccolithophores Emiliania huxleyi and Gephyrocapsa oceanica . Mar. Biotechnol. 1, 346–352 (1999)
Väänänen, H. K. & Parvinen, E. K. in The Carbonic Anhydrases (eds Tashian, R. E., Dodgson, S. J., Gros, G. & Carter, N. D. ) Ch. 32, 351–356 (Springer, 1991)
Kislyuk, A. O., Haegeman, B., Bergman, H. & Weitz, J. S. Genomic fluidity: an integrative view of gene diversity within microbial populations. BMC Genomics 12, 32 (2011)
Nanninga, H. J. & Tyrrell, T. Importance of light for the formation of algal blooms by Emiliania huxleyi . Mar. Ecol. Prog. Ser. 136, 195–203 (1996)
Havaux, M. et al. Vitamin B6 deficient plants display increased sensitivity to high light and photo-oxidative stress. BMC Plant Biol. 9, 130 (2009)
Zhu, S. H. & Green, B. R. Photoprotection in the diatom Thalassiosira pseudonana: role of LI818-like proteins in response to high light stress. Biochim. Biophys. Acta 1797, 1449–1457 (2010)
Xu, Y., Wahlund, T. M., Feng, L., Shaked, Y. & Morel, F. M. M. A novel alkaline phosphatase in the coccolithophore Emiliania huxleyi (Prymnesiophyceae) and its regulation by phosphorus. J. Phycol. 42, 835–844 (2006)
Karl, D. M. & Björkman, K. M. Dynamics of DOP in Biogeochemistry of Marine Dissolved Organic Matter (eds Hansell, D. A. & Carlson, C. A. ) Ch. 6, 249–348 (Elsevier Science, 2002)
Van Mooy, B. A. et al. Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458, 69–72 (2009)
Reid, E. L. et al. Coccolithophores: functional biodiversity, enzymes and bioprospecting. Mar. Drugs 9, 586–602 (2011)
Lessard, E. J., Merico, A. & Tyrell, T. Nitrate:phosphate ratios and Emiliania huxleyi blooms. Limnol. Oceanogr. 50, 1020–1024 (2005)
Turner, D. R., Hunter, K. A. & de Baar, H. J. W. The Biogeochemistry of Iron in Seawater Vol. 7, Ch. 1, 1–7 (John Wiley & Sons, 2001)
Erdner, D. L. & Anderson, D. M. Ferredoxin and flavodoxin as biochemical indicators of iron limitation during open-ocean iron enrichment. Limnol. Oceanogr. 44, 1609–1615 (1999)
Schulz, K. G. et al. Effect of trace metal availability on coccolithophorid calcification. Nature 430, 673–676 (2004)
Danbara, A. & Shiraiwa, Y. The requirement of selenium for the growth of marine coccolithophorids, Emiliania huxleyi, Gephyrocapsa oceanica and Helladosphaera sp. (Prymnesiophyceae). Plant Cell Physiol. 40, 762–766 (1999)
Sunda, W. G. & Huntsman, S. A. Feedback interactions between zinc and phytoplankton in seawater. Limnol. Oceanogr. 37, 25–40 (1992)
Acknowledgements
Joint Genome Institute (JGI) contributions were supported by the Office of Science of the US Department of Energy (DOE) under contract no. 7DE-AC02-05CH11231. We thank A. Gough for assistance with figures, C. Gentemann for Fig. 3 ocean colour analysis and P. Keeling for discussions.
Author information
Authors and Affiliations
Consortia
Contributions
Genome sequencing was performed by the US DOE JGI. B.A.R. coordinated the project and I.V.G. coordinated JGI sequencing/analysis; J.S. performed assemblies; A.K. and A.S. conducted automated annotation and analysis; U.J. at the AWI performed Illumina sequencing of 13 additional strains; A.K., X.Z., U.J., G.G., F.M., C.d.V., S.F., C.M., H.O., F.V., D.S., S.C.L., A.M., J.-M.C., Y.-C.L., Y.V.d.P., J.K., K.V., K.G., A.F.S., J.N., P.v.D. and G.W. performed genome and transcriptome analyses; U.J. and G.G. provided Illumina genomic sequence data, F.V. and D.S., tiling array data, and J. K., microarray data; J.Y. provided SEM images; phylogenetic anaylses was contributed by A.M., and A.Z.W. (Fig. 1b); E.K.H., M.J.K. and J.B.D. (Fig. 3c); J.M., C.F.D., M.A. U.J., and J.B.D (Supplementary Fig. 1); B.A.R. wrote the manuscript in collaboration with J.B.D., C.F.D., S.T.D., G.G., U.J., T.R., A.Z.W, X.Z. and I.V.G. (co-second senior authors). Authors in the first alphabetical list of the paper are equally contributing second authors who made substantial contributions to the paper. The remaining authors are members of the E. huxleyi Annotation Consortium who contributed additional analyses and/or annotations.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Text and Data in 5 sections – see contents for details. (PDF 3119 kb)
Supplementary Information
This file contains Supplementary Figures 1-6 and Supplementary Tables 1-15. (PDF 478 kb)
Supplementary Data 1
This file contains refined gene models validated by Sanger ESTs. (XLSX 765 kb)
Supplementary Data 2
This file contains refined gene models validated by tiling arrays. (XLSX 210 kb)
Supplementary Data 3
This file contains refined gene models validated by RNAseq. (XLSX 236 kb)
Supplementary Data 4
This file contains a phylogenomic gene list. (XLS 230 kb)
Supplementary Data 5
This file contains core and variable genes identified by direct mapping of Illumina reads based on 50% gene coverage. (XLSX 384 kb)
Supplementary Data 6
This file contains core genes identified by comparative genomic hybridization. (XLS 643 kb)
Supplementary Data 7
This file contains conserved eukaryotic genes mapping approach (CEGMA) list. (XLSX 64 kb)
Supplementary Data 8
This file contains genes encoding putative metal binding proteins. (XLSX 50 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-Non-Commercial-ShareAlike 3.0 Unported licence. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-sa/3.0/.
About this article
Cite this article
Read, B., Kegel, J., Klute, M. et al. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499, 209–213 (2013). https://doi.org/10.1038/nature12221
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12221
This article is cited by
-
Aerobic bacteria produce nitric oxide via denitrification and promote algal population collapse
The ISME Journal (2023)
-
A joint proteomic and genomic investigation provides insights into the mechanism of calcification in coccolithophores
Nature Communications (2023)
-
Effect of ocean acidification on the growth, response and hydrocarbon degradation of coccolithophore-bacterial communities exposed to crude oil
Scientific Reports (2023)
-
Abundant Sulfitobacter marine bacteria protect Emiliania huxleyi algae from pathogenic bacteria
ISME Communications (2023)
-
Rapid diversification underlying the global dominance of a cosmopolitan phytoplankton
The ISME Journal (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.