Abstract
The ever-increasing number of available microbial genomes and metagenomes provides new opportunities to investigate the links between niche partitioning and genome evolution in the ocean, especially for the abundant and ubiquitous marine picocyanobacteria Prochlorococcus and Synechococcus. Here, by combining metagenome analyses of the Tara Oceans dataset with comparative genomics, including phyletic patterns and genomic context of individual genes from 256 reference genomes, we show that picocyanobacterial communities thriving in different niches possess distinct gene repertoires. We also identify clusters of adjacent genes that display specific distribution patterns in the field (eCAGs) and are thus potentially involved in the same metabolic pathway and may have a key role in niche adaptation. Several eCAGs are likely involved in the uptake or incorporation of complex organic forms of nutrients, such as guanidine, cyanate, cyanide, pyrimidine, or phosphonates, which might be either directly used by cells, for example for the biosynthesis of proteins or DNA, or degraded to inorganic nitrogen and/or phosphorus forms. We also highlight the enrichment of eCAGs involved in polysaccharide capsule biosynthesis in Synechococcus populations thriving in both nitrogen- and phosphorus-depleted areas vs. low-iron (Fe) regions, suggesting that the complexes they encode may be too energy-consuming for picocyanobacteria thriving in the latter areas. In contrast, Prochlorococcus populations thriving in Fe-depleted areas specifically possess an alternative respiratory terminal oxidase, potentially involved in the reduction of Fe(III) to Fe(II). Altogether, this study provides insights into how phytoplankton communities populate oceanic ecosystems, which is relevant to understanding their capacity to respond to ongoing climate change.
Similar content being viewed by others
Introduction
Although phytoplankton communities play a crucial role in marine biogeochemical cycles [1, 2], the relative contribution of different species or ecotypes to these cycles remains difficult to assess due to a lack of knowledge of specific metabolic traits. Indeed, trait-based functional diversity is thought to be a better predictor of ecosystem functioning than species diversity [3, 4], and understanding which metabolic traits have facilitated the adaptation of an ecotype to a particular environment is key to understanding each species’ ecological role. Comparative genomics has sometimes been used to try to identify the genetic basis of niche adaptation. However, this approach has revealed only a few genes specific to particular ecotypes and thus potentially involved in niche adaptation, perhaps due to the fairly low number of genomes available (even for major phytoplankton groups) and the poor ecological representation and physiological characterization of available isolates [5,6,7,8,9]. An alternative to better deciphering the links between functional diversity and niche partitioning involves exploiting the rapidly growing number of metagenomes. These can be used to generate metagenome-assembled genomes (MAGs) that can fill the gaps for yet uncultured microbial taxa as well as to identify niche-specific genes, i.e. genes enriched in specific spatial and/or temporal environmental conditions, by recruiting metagenomic reads onto reference genomes [10,11,12,13,14,15,16].
Due to their abundance and ubiquity in the field and the large number of available genomes, including single amplified genomes (SAGs) and MAGs [6, 17,18,19,20], the marine picocyanobacteria Prochlorococcus and Synechococcus constitute highly pertinent models to study how phytoplankton cells adapt to their variable physico-chemical environment. These genera are indeed the two most abundant members of the phytoplankton community, Prochlorococcus being restricted to the 40°S-50°N latitudinal band, whilst the distribution of Synechococcus extends from the equator to subpolar waters [21,22,23]. By combining laboratory and environmental studies, scientists have managed to decipher their genetic diversity and delineate specific ecotypes or “ecologically significant taxonomic units” (ESTUs), i.e. genetic groups within clades occupying a specific ecological niche [24,25,26,27,28]. While three major ESTU assemblages were identified for Prochlorococcus in surface waters, whose distribution was found to be mainly driven by temperature and iron (Fe) availability, eight distinct assemblages were identified for Synechococcus depending on three main environmental parameters: temperature, Fe, and phosphorus (P) availability. Nevertheless, few studies have so far linked knowledge of the distribution of the different ecotypes to their functional diversity in order to identify potential niche-specific genes, based on gene relative abundance in different ecosystems [10, 14, 15, 29, 30]. Furthermore, most of these previous studies have focused on the abundance of individual genes, or more rarely, on just a few genomic regions with known function, for example those involved in nitrogen (N) or P uptake and assimilation [31,32,33].
Here, in order to better understand the relationship between biogeochemistry and metabolic traits of marine picocyanobacteria, we searched for global patterns of picocyanobacterial gene distributions. To do so, we used a network approach to integrate metagenome analyses of the oceanwide Tara Oceans dataset and synteny of individual accessory genes in 256 reference genomes, SAGs, or MAGs covering the wide diversity existing within Prochlorococcus and Synechococcus. Using this approach, we identified many clusters of adjacent genes that display distinctive global distribution patterns in situ and thus likely play important roles in the adaptation of these bacteria to the main ecological niches they occupy in the ocean. Given that gene synteny is commonly used as an indicator of shared function [34, 35], delineation of these gene clusters should also help to identify the putative function of numerous unknown genes, based on their occurrence alongside functionally annotated genes in the same cluster. Overall, this study provides novel insights into the genetic basis of niche partitioning in key members of the phytoplankton community.
Results and discussion
Different picocyanobacterial communities exhibit distinct gene repertoires
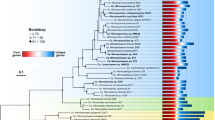
To analyze the distribution of Prochlorococcus and Synechococcus reads along the Tara Oceans transect, metagenomic reads corresponding to the bacterial size fraction were recruited against 256 picocyanobacterial reference genomes, including SAGs and MAGs representative of uncultured lineages (e.g., Prochlorococcus HLIII-IV, Synechococcus EnvA or EnvB). This yielded a total of 1.07 billion recruited reads, of which 87.7% mapped onto Prochlorococcus genomes and 12.3% onto Synechococcus genomes, which were then functionally assigned by mapping them onto the manually curated Cyanorak v2.1 CLOG database [19]. In order to identify picocyanobacterial genes potentially involved in niche adaptation, we analyzed the distribution across the oceans of flexible (i.e. non-core) genes. Clustering of Tara Oceans stations according to the relative abundance of flexible genes resulted in three well-defined clusters for Prochlorococcus (Fig. 1A), which matched those obtained when stations were clustered according to the relative abundance of Prochlorococcus ESTUs, as assessed using the high-resolution marker gene petB, encoding cytochrome b6 (Fig. 1A; [24]). Only a few discrepancies can be observed between the two trees, including stations TARA-070 that displayed one of the most disparate ESTU compositions and TARA-094, dominated by the rare HLID ESTU (Fig. 1A). Similarly, for Synechococcus, most of the eight assemblages of stations discriminated based on the relative abundance of ESTUs (Fig. 1B) were also retrieved in the clustering based on flexible gene abundance, except for a few intra-assemblage switches between stations, notably those dominated by ESTU IIA (Fig. 1B). Despite these few variations, four major clusters can be clearly delineated in both Synechococcus trees, corresponding to four broadly defined ecological niches, namely (i) cold, nutrient-rich, pelagic or coastal environments (blue and light red in Fig. 1B), (ii) Fe-limited environments (purple and grey), (iii) temperate, P-depleted, Fe-replete areas (yellow) and (iv) warm, N-depleted, Fe-replete regions (dark red). This correspondence between taxonomic and functional information was also confirmed by the high congruence between distance matrices based on ESTU relative abundance and on CLOG relative abundance (p-value < 10−4, mantel test r = 0.84 and r = 0.75 for Synechococcus and Prochlorococcus, respectively; dataset 1–4). Altogether, this indicates that distinct picocyanobacterial communities, as assessed based on a single taxonomic marker, also display different gene repertoires. As previously suggested for Prochlorococcus [36], this strong correlation between taxonomy and gene content strengthens the idea that, in both genera, the evolution of the accessory genome mainly occurs by vertical transmission, with a relatively low extent of lateral gene transfers, although we cannot exclude that the latter events may occur more often within members of a given ecotype.
A Prochlorococcus. B Synechococcus. Leaves of the trees correspond to stations along the Tara Oceans transect that are colored according to the code shown at the bottom of the trees, which corresponds to ESTU assemblages as determined previously [24] by clustering stations exhibiting similar ESTU relative abundance profiles shown here on the right of each tree (for global distribution maps of ESTU assemblages, see Figs. 3B and 4B in [24]). ESTUs were colored according to the palette below each panel. Dotted lines in dendrograms indicate discrepancies between tree topologies. Accessory genes correspond to all genes except those defined as large-core genes in a previous study [6]. Of note, due to a slightly different clustering method (cf. materials and methods), assemblage 7 (dark grey stations in 1B), which was discriminated from assemblage 6 in the Farrant et al. (2016) now clusters with this assemblage. Abbreviations: IO Indian Ocean, MS Mediterranean Sea, NAO North Atlantic Ocean, NPO North Pacific Ocean, RS Red Sea, SAO South Atlantic Ocean, SO Southern Ocean.
Distribution of flexible genes is tightly linked to environmental parameters and ESTUs
In order to reduce the amount of data and better interpret the global distribution of picocyanobacterial gene content, a correlation network of genes was built for each genus based on relative abundance profiles of genes across Tara Oceans samples using Weighted Correlation Network Analysis (WGCNA). This analysis emphasized four main modules of genes for Prochlorococcus (Fig. S1A) and five for Synechococcus (Fig. S1B), each module being abundant in a different set of stations. These modules were then associated with the available environmental parameters (Fig. 2A, B) and to the relative abundance of Prochlorococcus or Synechococcus ESTUs at each station (Fig. 2C, D). For instance, the Prochlorococcus brown module was strongly correlated with nutrient concentrations, particularly nitrate and phosphate, and strongly anti-correlated with Fe availability (Fig. 2A). This module thus corresponds to genes preferentially found in Fe-limited high-nutrient low-chlorophyll (HNLC) areas. Indeed, the brown module eigengene (Fig. S1A), i.e. the first principal component of gene abundances at the different stations for this module, representative of the abundance profiles of genes for this module at the different stations, showed the highest abundances at stations TARA-100 to 125, localized in the South and North Pacific Ocean, as well as at TARA-052, a station located close to the northern coast of Madagascar, likely influenced by the Indonesian throughflow originating from the tropical Pacific Ocean [24, 37]. Furthermore, the correlation of the Prochlorococcus brown module with the relative abundance of ESTUs at each station showed that it is also strongly associated with the presence of HLIIIA and HLIVA (Fig. 2C), previously shown to constitute the dominant Prochlorococcus ESTUs in low-Fe environments [24, 38, 39] but also the LLIB ESTU, found to dominate the LLI population in these low-Fe areas [24]. Altogether, this example and analyses of all other Prochlorococcus and Synechococcus modules (supplementary text) show that the communities colonizing cold, Fe-, N-, and/or P-depleted niches possess specific gene repertoires potentially involved in their adaptation to these particular environmental conditions.
A, B Correlation of module eigengenes to physico-chemical parameters for Prochlorococcus (A) and Synechococcus (B). C, D Correlation of module eigengenes to relative abundance profiles of ESTUs sensu [4]. Pearson (A, B) and Spearman (C, D) correlation coefficients (r and rho, respectively) are indicated by the color scale. Each module is identified by a specific color and the number between brackets specifies the number of genes in each module. The eigengene is representative of the relative abundance of genes of a given module at each Tara Oceans station. Non-significant correlations (Student asymptotic p-value > 0.01) are marked by a cross. Φsat: index of iron limitation derived from satellite data. PAR30: satellite-derived photosynthetically available radiation at the surface, averaged on 30 days. DCM: depth of the deep chlorophyll maximum.
Identification of individual genes potentially involved in niche partitioning
To identify genes relevant for adaptation to a specific set of environmental conditions and enriched in specific ESTU assemblages, we selected the most representative genes from each module (Dataset 5; Figs. 3, S2). Most genes retrieved this way encode proteins of unknown or hypothetical function (85.7% of 7,485 genes). However, among the genes with a functional annotation (Dataset 6), a large fraction seems to have a function related to their realized environmental niche (Figs. 3, S2). For instance, many genes involved in the transport and assimilation of nitrite and nitrate (nirA, nirX, moaA-C, moaE, mobA, moeA, narB, M, nrtP; [6]) as well as cyanate, an organic form of nitrogen (cynA, B, D, S), are enriched in the Prochlorococcus blue module, which is correlated with the HLIIA-D ESTU and to low inorganic N, P, and silica levels and anti-correlated with Fe availability (Fig. 2A–C). This is consistent with previous studies showing that while only a few Prochlorococcus strains in culture possess the nirA gene and even less the narB gene, natural Prochlorococcus populations inhabiting N-poor areas do possess one or both of these genes [40,41,42]. Similarly, numerous genes amongst the most representative of Prochlorococcus brown, red and turquoise modules are related to adaptation of HLIIIA/IVA, HLIA and LLIA ESTUs to Fe-limited, cold P-limited, and cold, mixed waters, respectively (Fig. 3). Comparable results were obtained for Synechococcus, although the niche delineation was less clear than for Prochlorococcus since genes within each module exhibited lower correlations with the module eigenvalue (Fig. S2). These results therefore constitute a proof of concept that this network analysis was able to retrieve niche-related genes from metagenomics data.
For each module, each gene is represented as a dot positioned according to its correlation with the eigengene for each module, the most representative genes being localized on top of each violin plot. Genes mentioned in the text and/or in Dataset 6 have been colored according to the color of the corresponding module, indicated by a colored bar above each module. The text above violin plots indicates the most significant environmental parameter(s) and/or ESTU(s) for each module, as derived from Fig. 2.
Identification of eCAGs potentially involved in niche partitioning
In order to better understand the function of niche-related genes, notably of the numerous unknowns, we then integrated global distribution data with gene synteny in reference genomes using a network approach (Datasets 7, 8). This led us to identify clusters of adjacent genes in reference genomes, and thus potentially involved in the same metabolic pathway (Figs. 4, S3, S4; Dataset 6). These clusters were defined within each module and thus encompass genes with similar distribution and abundance in situ. Hereafter, these environmental clusters of adjacent genes will be called “eCAGs”.
Nodes correspond to individual genes with their gene name (or significant numbers of the CK number, e.g. 1234 for CK_00001234) and are colored according to their WGCNA module. A link between two nodes indicates that these two genes are less than five genes apart in at least one genome. The bottom insert shows the most significant environmental parameter(s) and/or ESTU(s) for each module, as derived from Fig. 2.
eCAGs related to nitrogen metabolism
The well-known nitrate/nitrite gene cluster involved in uptake and assimilation of inorganic forms of N (see above), which is present in most Synechococcus genomes (Dataset 6), was expectedly not restricted to a particular niche in natural Synechococcus populations, as shown by its quasi-absence from WGCNA modules. In Prochlorococcus, this cluster is separated into two eCAGs enriched in low-N areas (Fig. S5A, B), most genes being included in Pro-eCAG_002, present in only 13 out of 118 Prochlorococcus genomes, while nirA and nirX form an independent eCAG (Pro-eCAG_001) due to their presence in many more genomes. The quasi-core ureA-G/urtB-E genomic region was also found to form a Prochlorococcus eCAG (Pro-eCAG_003) that was impoverished in low-Fe compared to other regions (Fig. S5C, D), in agreement with its presence in only two out of six HLIII/IV genomes. We also uncovered several other Prochlorococcus and Synechococcus eCAGs that seem to be involved in the transport and/or assimilation of more unusual and/or complex forms of nitrogen, which might either be degraded into elementary N molecules or possibly directly used by cells for e.g. the biosynthesis of proteins or DNA. Indeed, we detected in both genera an eCAG (Pro-eCAG_004 and Syn-eCAG_001; Fig. S6A, B; Dataset 6) that encompasses speB2, an ortholog of Synechocystis PCC 6803 sll1077, previously annotated as encoding an agmatinase [29, 43] and which was recently characterized as a guanidinase that degrades guanidine rather than agmatine to urea and ammonium [44]. E. coli produces guanidine under nutrient-poor conditions, suggesting that guanidine metabolism is biologically significant and potentially prevalent in natural environments [44, 45]. Furthermore, the ykkC riboswitch candidate, which was shown to specifically sense guanidine and to control the expression of a variety of genes involved in either guanidine metabolism or nitrate, sulfate, or bicarbonate transport, is located immediately upstream of this eCAG in Synechococcus reference genomes, all genes of this cluster being predicted by RegPrecise 3.0 to be regulated by this riboswitch (Fig. S6C; [45, 46]). The presence of hypA and B homologs within this eCAG furthermore suggests that, in the presence of guanidine, these homologs could be involved in the insertion of Ni2+, or another metal cofactor, in the active site of guanidinase. The next three genes of this eCAG, which encode an ABC transporter similar to the TauABC taurine transporter in E. coli (Fig. S6C), could be involved in guanidine transport in low-N areas. Of note, the presence in most Synechococcus/Cyanobium genomes possessing this eCAG of a gene encoding a putative Rieske Fe-sulfur protein (CK_00002251) downstream of this gene cluster, seems to constitute a specificity compared to the homologous gene cluster in Synechocystis sp. PCC 6803. The presence of this Fe-S protein suggests that Fe is used as a cofactor in this system and might explain why this gene cluster is absent from picocyanobacteria thriving in low-Fe areas, while it is present in a large proportion of the population in most other oceanic areas (Fig. S6A, B).
Another example of the use of organic N forms concerns compounds containing a cyano radical (C ≡ N). The cyanate transporter genes (cynABD) were indeed found in a Prochlorococcus eCAG (Pro-eCAG_005, also including the conserved hypothetical gene CK_00055128; Fig. S7A, B). While only a small proportion of the Prochlorococcus community possesses this eCAG in warm, Fe-replete waters, it is absent from other oceanic areas in accordance with its low frequency in Prochlorococcus genomes (present in only two HLI and five HLII genomes). In Synechococcus these genes were not included in a module, and thus are not in an eCAG (Dataset 6; Fig. S7C), but seem widely distributed despite their presence in only a few Synechococcus genomes (mostly in clade III strains; [6, 47, 48]). Interestingly, we also uncovered a 7-gene eCAG (Pro-eCAG_006 and Syn-eCAG_002), encompassing a putative nitrilase gene (nitC), which also suggests that most Synechococcus cells and a more variable fraction of the Prochlorococcus population could use nitriles or cyanides in warm, Fe-replete waters and more particularly in low-N areas such as the Indian Ocean (Fig. 5A, B). The whole operon (nitHBCDEFG; Fig. 5C), called Nit1C, was shown to be upregulated in the presence of cyanide and to trigger an increase in the rate of ammonia accumulation in the heterotrophic bacterium Pseudomonas fluorescens [49], suggesting that like cyanate, cyanide could constitute an alternative nitrogen source in marine picocyanobacteria as well. However, given the potential toxicity of these C ≡ N-containing compounds [50], we cannot exclude that these eCAGs could also be devoted to cell detoxification [45, 47]. Such an example of detoxification has been described for arsenate and chromate that, as analogs of phosphate and sulfate respectively, are toxic to marine phytoplankton and must be actively exported out of the cells [51, 52].
A Prochlorococcus Pro-eCAG_006. B Synechococcus Syn-eCAG_002. C The genomic region in Prochlorococcus marinus MIT9301. The size of the circle is proportional to relative abundance of each genus as estimated based on the single-copy core gene petB and this gene was also used to estimate the relative abundance of other genes in the population. Black dots represent Tara Oceans stations for which Prochlorococcus or Synechococcus read abundance was too low to reach the threshold limit.
We detected the presence of an eCAG encompassing asnB, pyrB2, and pydC (Pro-eCAG_007, Syn-eCAG_003, Fig. S8), which could contribute to an alternative pyrimidine biosynthesis pathway and thus provide another way for cells to recycle complex nitrogen forms. While this eCAG is found in only one fifth of HLII genomes and in quite specific locations for Prochlorococcus, notably in the Red Sea, it is found in most Synechococcus cells in warm, Fe-replete, N and P-depleted niches, consistent with its phyletic pattern showing its absence only from most clade I, IV, CRD1, and EnvB genomes (Fig. S8; Dataset 6). More generally, most N-uptake and assimilation genes in both genera were specifically absent from Fe-depleted areas, including the nirA/narB eCAG for Prochlorococcus, as mentioned by Kent et al. [36] as well as guanidinase and nitrilase eCAGs. In contrast, picocyanobacterial populations present in low-Fe areas possess, in addition to the core ammonium transporter amt1, a second transporter amt2, also present in cold areas for Synechococcus (Fig. S9). Additionally, Prochlorococcus populations thriving in HNLC areas also possess two amino acid-related eCAGs that are present in most Synechococcus genomes, the first one involved in polar amino acid N-II transport (Pro-eCAG_008; natF-G-H-bgtA; [53]; Fig. S10A, B) and the second one (leuDH-soxA-CK_00001744, Pro-eCAG_009, Fig. S10C, D) that notably encompasses a leucine dehydrogenase, able to produce ammonium from branched-chain amino acids. This highlights the profound difference in N acquisition mechanisms between HNLC regions and Fe-replete, N-deprived areas: the primary nitrogen sources for picocyanobacterial populations dwelling in HNLC areas seem to be ammonium and amino acids, while N acquisition mechanisms are more diverse in N-limited, Fe-replete regions.
eCAGs related to phosphorus metabolism
Adaptation to P depletion has been well documented in marine picocyanobacteria showing that while in P-replete waters Prochlorococcus and Synechococcus essentially rely on inorganic phosphate acquired by core transporters (PstSABC), strains isolated from low-P regions and natural populations thriving in these areas additionally contain a number of accessory genes related to P metabolism, located in specific genomic islands [6, 14, 30,31,32, 54]. Here, we indeed found in Prochlorococcus an eCAG containing the phoBR operon (Pro-eCAG_010) that encodes a two-component system response regulator, as well as an eCAG including the alkaline phosphatase phoA (Pro-eCAG_011), both present in virtually the whole Prochlorococcus population from the Mediterranean Sea, the Gulf of Mexico and the Western North Atlantic Ocean, which are known to be P-limited [30, 55] (Fig. S11A, B). By comparison, in Synechococcus, we only identified the phoBR eCAG (Syn-eCAG_005, Fig. S11C) that is systematically present in warm waters whatever the limiting nutrient, in agreement with its phyletic pattern in reference genomes showing its specific absence from cold thermotypes (clades I and IV, Dataset 6). Furthermore, although our analysis did not retrieve them within eCAGs due to the variability of gene content and synteny in this genomic region, even within each genus, several other P-related genes were enriched in low-P areas but partially differed between Prochlorococcus and Synechococcus (Figs. 3, S2, S11; Dataset 6). While the genes putatively encoding a chromate transporter (ChrA) and an arsenate efflux pump ArsB were present in both genera in different proportions, a putative transcriptional phosphate regulator related to PtrA (CK_00056804; [56]) was specific to Prochlorococcus. Synechococcus in contrast harbors a large variety of alkaline phosphatases (PhoX, CK_00005263 and CK_00040198) as well as the phosphate transporter SphX (Fig. S11).
Phosphonates, i.e. reduced organophosphorus compounds containing C–P bonds that represent up to 25% of the high-molecular-weight dissolved organic P pool in the open ocean, constitute an alternative P form for marine picocyanobacteria [57]. We indeed identified, in addition to the core phosphonate ABC transporter (phnD1-C1-E1), a second previously unreported putative phosphonate transporter phnC2-D2-E2-E3 (Pro-eCAG_012; Fig. 6A). Most of the Prochlorococcus population in strongly P-limited areas of the ocean harbored these genes, while they were absent from other areas, consistent with their presence in only a few Prochlorococcus and no Synechococcus genomes. Furthermore, as previously described [58,59,60], we found a Prochlorococcus eCAG encompassing the phnYZ operon involved in C-P bond cleavage, the putative phosphite dehydrogenase ptxD, and the phosphite and methylphosphonate transporter ptxABC (Pro-eCAG_0013, Dataset 6; Fig. 6B, [60,61,62]). Compared to these previous studies that mainly reported the presence of these genes in Prochlorococcus cells from the North Atlantic Ocean, here we show that they actually occur in a much larger geographic area, including the Mediterranean Sea, the Gulf of Mexico, and the ALOHA station (TARA_132) in the North Pacific, even though they were present in a fairly low fraction of Prochlorococcus cells. These genes occurred in an even larger proportion of the Synechococcus population, although not found in an eCAG for this genus (Fig. S12; Dataset 6). Synechococcus cells from the Mediterranean Sea, a P-limited area dominated by clade III [24], seem to lack phnYZ, in agreement with the phyletic pattern of these genes in reference genomes, showing the absence of this two-gene operon in the sole clade III strain that possesses the ptxABDC gene cluster. In contrast, the presence of the complete gene set (ptxABDC-phnYZ) in the North Atlantic, at the entrance of the Mediterranean Sea, and in several clade II reference genomes rather suggests that it is primarily attributable to this clade. Altogether, our data indicate that part of the natural populations of both Prochlorococcus and Synechococcus would be able to assimilate phosphonate and phosphite as alternative P-sources in low-P areas using the ptxABDC-phnYZ operon. Yet, the fact that no picocyanobacterial genome except P. marinus RS01 (Fig. 6C) possesses both phnC2-D2-E2-E3 and phnYZ, suggests that the phosphonate taken up by the phnC2-D2-E2-E3 transporter could be incorporated into cell surface phosphonoglycoproteins that may act to mitigate cell mortality by grazing and viral lysis, as recently suggested [63].
A Prochlorococcus Pro-eCAG_012 putatively involved in phosphonate transport. B Prochlorococcus Pro-eCAG_013, involved in phosphonate/phosphite uptake and assimilation and phosphonate C-P bond cleavage. C The genomic region encompassing both phnC2-D2-E2-E3 and ptxABDC-phnYZ specific to P. marinus RS01. The size of the circle is proportional to relative abundance of Prochlorococcus as estimated based on the single-copy core gene petB and this gene was also used to estimate the relative abundance of other genes in the population. Black dots represent Tara Oceans stations for which Prochlorococcus read abundance was too low to reach the threshold limit.
eCAGs related to iron metabolism
As for macronutrients, it has been hypothesized that the survival of marine picocyanobacteria in low-Fe regions was made possible through several strategies, including the loss of genes encoding proteins that contain Fe as a cofactor, the replacement of Fe by another metal cofactor, and the acquisition of genes involved in Fe uptake and storage [14, 15, 36, 39, 64]. Accordingly, several eCAGs encompassing genes encoding proteins interacting with Fe were found in modules anti-correlated to HNLC regions in both genera. These include three subunits of the (photo)respiratory complex succinate dehydrogenase (SdhABC, Pro-eCAG_014, Syn-eCAG_006, Fig. S13; [65]) and Fe-containing proteins encoded in most abovementioned eCAGs involved in N or P metabolism, such as the guanidinase (Fig. S6), the NitC1 (Fig. 5), the pyrB2 (Fig. S8), the phosphonate (Fig. 6, S12), and the urea and inorganic nitrogen eCAGs (Fig. S5). Most Synechococcus cells thriving in Fe-replete areas also possess the sodT/sodX eCAG (Syn-eCAG_007, Fig. S14A, B) involved in nickel transport and maturation of the Ni-superoxide dismutase (SodN), these three genes being in contrast core in Prochlorococcus. Additionally, Synechococcus from Fe-replete areas, notably from the Mediterranean Sea and the Indian Ocean, specifically possess two eCAGs (Syn-eCAG_008 and 009; Fig. S14C, D), involved in the biosynthesis of a polysaccharide capsule that appear to be most similar to the E. coli groups 2 and 3 kps loci [66]. These extracellular structures, known to provide protection against biotic or abiotic stress, were recently shown in Klebsiella to provide a clear fitness advantage in nutrient-poor conditions since they were associated with increased growth rates and population yields [67]. However, while these authors suggested that capsules may play a role in Fe uptake, the significant reduction in the relative abundance of kps genes in low-Fe compared to Fe-replete areas (t-test p-value < 0.05 for all genes of the Syn-eCAG_008 and 009; Fig. S14C) and their absence in CRD1 strains (Dataset 6) rather suggests that these capsules may be too energy-consuming for some picocyanobacteria thriving in this particular niche, while they may have a more meaningful and previously overlooked role in their adaptation to low-P and low-N niches.
Several eCAGs were in contrast enriched in populations dwelling in HNLC environments, dominated by Prochlorococcus HLIIIA/HLIVA/LLIB and Synechococcus CRD1A/EnvBA ESTUs (Fig. 2). The vast majority of Prochlorococcus cells thriving in low-Fe regions possess an eCAG encompassing the ctaC2-D2-E2 operon, also found in 85% of all Synechococcus reference genomes, including all CRD1 (Fig. 7; Dataset 6). This eCAG encodes the alternative respiratory terminal oxidase ARTO, a protein complex that has been suggested to be part of a minimal respiratory chain in the cytoplasmic membrane, potentially providing an additional electron sink under Fe-deprived conditions [68, 69]. Furthermore, a Synechocystis mutant in which the ctaD2 and ctaE2 genes were inactivated was found to display markedly impaired Fe reduction and uptake rates as compared to wild-type cells, suggesting that ARTO is involved in the reduction of Fe(III) to Fe(II) prior to its transport through the plasma membrane via the Fe(II) transporter FeoB [70]. Thus, the presence of the ARTO system appears to represent a major and previously unreported adaptation for Prochlorococcus populations thriving in low-Fe areas.
A Prochlorococcus Pro-eCAG_016. B Synechococcus Syn-eCAG_015. C The genomic region encompassing the ctaC2D2E2 operon in P. marinus MIT9201. The size of the circle is proportional to relative abundance of each genus as estimated based on the single-copy core gene petB and this gene was also used to estimate the relative abundance of other genes in the population. Black dots represent Tara Oceans stations for which Prochlorococcus or Synechococcus read abundance was too low to reach the threshold limit.
Both Prochlorococcus and Synechococcus thriving in low-Fe waters also possess eCAGs encoding the TonB-dependent siderophore uptake operon fecDCAB-tonB-exbBD (Pro-eCAG_015 and Syn-eCAG_013-014, Dataset 6). This gene cluster, which is found in a few picocyanobacterial genomes and was previously shown to be anti-correlated with dissolved Fe concentration [14, 15, 64], is indeed systematically present in a significant part of the Prochlorococcus and Synechococcus population in low-Fe areas (Fig. S15). However, it is also present in a small fraction of the populations thriving in the Indian Ocean, consistent with its occurrence in two Prochlorococcus HLII and one Synechococcus clade II genomes (Dataset 6). Finally, a large proportion of the Synechococcus populations in HNLC areas possess (i) a large eCAG involved in glycine betaine synthesis and transport (Syn-eCAG_010, Fig. S15A, B; [6, 71, 72]), almost absent from low-N areas, (ii) an eCAG encompassing a flavodoxin and a thioredoxin reductase (Syn-eCAG_011, Fig. S16C, D), mostly absent from low-P areas, and (iii) the nfeD-floT1-floT2 eCAG (Syn-eCAG_012, Fig. S17A, B) involved in the production of lipid rafts, potentially affecting cell shape and motility [6, 73].
eCAGs enriched in cold waters
Besides genes involved in nutrient acquisition and metabolism, several Prochlorococcus and Synechococcus eCAGs were found to be correlated with low temperature. A closer examination of Prochlorococcus eCAGs however, shows that their occurrence is not directly related to temperature adaptation but mainly explained by the prevalence at high latitude of either (i) the HLIA ESTU (Figs. 2A, C, 4), the red module encompassing most of the above-mentioned eCAGs involved in P-uptake and assimilation pathways, or (ii) the LLIA ESTU, present in surface waters at vertically-mixed stations, the turquoise module mainly gathering Prochlorococcus LL-specific genes, such as Pro-eCAG_017, involved in phycoerythrin-III biosynthesis (ppeA, cpeFTZY, unk13). As concerns Synechococcus, although a fairly high number of eCAGs were identified in the tan module associated with ESTUs IA and IVA-C (Fig. 2B, D and Fig. S4), only very few are conserved in more than two reference strains and/or have a characterized function (Dataset 6). Among these, at least one eCAG is clearly related to adaptation to cold waters, the orange carotenoid protein (OCP) operon (ocp-crtW-frp; Syn-eCAG_016). Indeed, this operon is involved in a photoprotective process [74] that provides cells with the ability to deal with oxidative stress under cold temperatures [75]. Accordingly, our data shows that Synechococcus populations colonizing mixed waters at high latitudes or in upwelling areas all possess this eCAG (Fig. S18), highlighting the importance of this photoprotection system in Synechococcus populations colonizing cold and temperate areas. Synechococcus populations thriving in cold waters also appear to be enriched in eCAGs involved in gene regulation such as transcription factors involved in the regulation of the CA4-A form of the type IV chromatic acclimation process (fciA-B; Syn-eCAG_017), consistent with the predominance of Synechococcus CA4-A cells in temperate or cold environments [76,77,78] (Dataset 6). Altogether, the fairly low number of eCAGs strongly associated with temperature supports the hypothesis that adaptation to cold temperature is not mediated by evolution of gene content but rather of protein sequences [5, 6, 36, 79].
Conclusions
Our analysis of Prochlorococcus and Synechococcus gene distributions at the global scale using the deeply sequenced metagenomes collected along the Tara Oceans expedition transect revealed that each picocyanobacterial community has a specific gene repertoire, with different sets of accessory genes being highly correlated with distinct ESTUs and physicochemical parameters. As previously suggested for Prochlorococcus [36], this strong correlation between taxonomy and gene content strengthens the idea that, in both genera, genome evolution mainly occurs by vertical transmission and selective gene retention, and that lateral gene transfers between ecotypes are fairly scarce. By combining information about gene synteny in 256 reference genomes with the distribution and abundance of these genes in the field, we further managed to delineate suites of adjacent genes likely involved in the same metabolic pathways that may have a crucial role in adaptation to specific niches. These analyses confirmed previous observations about the niche partitioning of individual genes and a few genomic regions involved in nutrient uptake and assimilation [14, 15, 25, 31, 33, 36, 40, 42]. Most importantly, even though our network approach likely only revealed the lower boundary of the number of eCAGs actually present in different niches, due to the incompleteness of some reference genomes, this approach highlighted that some previously detected individual genes are part of larger genomic regions and unveiled several novel genomic regions. Although we cannot exclude that some genes enriched in a specific niche are not adaptive per se but either hitchhiked along with an adaptive gene [80] or occurred from passive transport of ecotype populations outside their niche [79, 81], it is reasonable to assume that many eCAGs identified in the present study could confer cells a fitness benefit in particular niches and were thus retained by natural selection, or in contrast have been counter-selected in certain environments (such as eCAGs that are absent from low-Fe environments). This study revealed the potential importance of the uptake and assimilation of organic forms of nutrients, which might either be directly used by cells e.g. for the biosynthesis of proteins or DNA, or be degraded into inorganic N and/or P forms. Consequently, many eCAGs potentially involved in the uptake and assimilation of complex compounds, such as guanidine, C ≡ N-containing compounds, or pyrimidine were present in both N- and P-depleted waters, and might constitute an advantage in areas of the world ocean co-limited in these nutrients [30], while they were absent from HNLC areas. Our data also suggests that adaptation to Fe-limitation relies on specific adaptation mechanisms including the reduction of Fe(III) to Fe(II) using ARTO, Fe scavenging using siderophores, as well as reduction of the Fe quota and of energy-consuming mechanisms, such as polysaccharide capsule biosynthesis. Altogether, this study provides novel insights into the genetic basis of niche partitioning in key members of the phytoplankton community. A future challenge will clearly be biochemically characterizing the function of adaptive genes in these eCAGs (Datasets 5, 6), which are sometimes present in only a few or even a single cultured strain but which can occur in a large part or even the whole Prochlorococcus and/or Synechococcus population occupying a specific niche in situ.
Materials and methods
Tara Oceans metagenomic reads from surface waters corresponding to the bacterial size-fraction [24, 82] were recruited against 256 reference genomes using MMseqs2 Release 11-e1a1c (76) and then mapped to an extended database, including 722 outgroup cyanobacterial genomes (Dataset 9; Supplementary Methods). Picocyanobacterial reads were then taxonomically assigned to either Prochlorococcus or Synechococcus and functionally assigned to a cluster of likely orthologous genes (CLOGs) as defined in the information system Cyanorak v2.1 [19]. After normalization by gene and read length, filtration steps included the selection of (i) samples containing more CLOGs than the average number of genes in a Synechococcus or Prochlorococcus HL genome, (ii) CLOGs with a gene coverage higher than 1 in at least 2 of the selected samples and (iii) non-core genes [6].
Tara Oceans stations were clustered using Ward’s minimum variance [83] based on Bray-Curtis similarities between the relative abundance of either CLOG or picocyanobacterial ESTUs as defined based on the petB marker gene [24]. CLOG abundance profiles were also used to perform co-occurrence analyses by weighted genes correlation network analysis (WGCNA, [84, 85]) to delineate modules of CLOGs that share a similar distribution pattern. The eigengene of each module was then correlated to environmental parameters, retrieved from PANGAEA (www.pangaea.de/), and to the relative abundance of petB ESTUs. Furthermore, the most representative genes of each module were identified as those most correlated to the eigengene.
Finally, we then defined eCAGs within each module by searching adjacent genes (less than 6 genes apart in 80% of the genomes possessing them) in the 256 reference genomes (Datasets 7, 8) that were used to build a network of the corresponding CLOGs (node) according to the graph embedder (GEM) or the Fruchterman-Reingold layout algorithms implemented in the R package igraph [86].
Data availability
All genome sequences used in this study are available from NCBI as detailed in Dataset 9, while Tara Oceans metagenomes and corresponding environmental parameters were retrieved from PANGAEA (www.pangaea.de/).
References
Field CB, Behrenfeld MJ, Randerson JT, Falkowski PG. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998;281:237–40.
Litchman E, de Tezanos Pinto P, Edwards KF, Klausmeier CA, Kremer CT, Thomas MK. Global biogeochemical impacts of phytoplankton: a trait-based perspective. J Ecol. 2015;103:1384–96.
Abonyi A, Horváth Z, Ptacnik R. Functional richness outperforms taxonomic richness in predicting ecosystem functioning in natural phytoplankton communities. Freshw Biol. 2018;63:178–86.
Ye L, Chang C-W, Matsuzaki SS, Takamura N, Widdicombe CE, Hsieh C. Functional diversity promotes phytoplankton resource use efficiency. J Ecol. 2019;107:2353–63.
Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231.
Doré H, Farrant GK, Guyet U, Haguait J, Humily F, Ratin M, et al. Evolutionary mechanisms of long-term genome diversification associated with niche partitioning in marine picocyanobacteria. Front Microbiol. 2020;11:567431.
Nef C, Madoui M-A, Pelletier É, Bowler C. Whole-genome scanning reveals environmental selection mechanisms that shape diversity in populations of the epipelagic diatom Chaetoceros. PLoS Biol. 2022;20:e3001893.
Read BA, Kegel J, Klute MJ, Kuo A, Lefebvre SC, Maumus F, et al. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature. 2013;499:209–13.
Piganeau G, Grimsley N, Moreau H. Genome diversity in the smallest marine photosynthetic eukaryotes. Res Microbiol. 2011;162:570–7.
Delmont TO, Eren AM. Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ. 2018;6:e4320.
Tully BJ, Graham ED, Heidelberg JF. The reconstruction of 2631 draft metagenome-assembled genomes from the global oceans. Sci Data. 2018;5:170203.
Massana R, López-Escardó D. Metagenome assembled genomes are for eukaryotes too. Cell Genom. 2022;2:100130.
Duncan A, Barry K, Daum C, Eloe-Fadrosh E, Roux S, Schmidt K, et al. Metagenome-assembled genomes of phytoplankton microbiomes from the Arctic and Atlantic Oceans. Microbiome. 2022;10:67.
Garcia CA, Hagstrom GI, Larkin AA, Ustick LJ, Levin SA, Lomas MW, et al. Linking regional shifts in microbial genome adaptation with surface ocean biogeochemistry. Philos Trans R Soc B Biol Sci. 2020;375:20190254.
Ahlgren NA, Belisle BS, Lee MD. Genomic mosaicism underlies the adaptation of marine Synechococcus ecotypes to distinct oceanic iron niches. Environ Microbiol. 2020;22:1801–15.
Garcia SL, Stevens SLR, Crary B, Martinez-Garcia M, Stepanauskas R, Woyke T, et al. Contrasting patterns of genome-level diversity across distinct co-occurring bacterial populations. ISME J. 2018;12:742–55.
Berube PM, Biller SJ, Hackl T, Hogle SL, Satinsky BM, Becker JW, et al. Single cell genomes of Prochlorococcus, Synechococcus, and sympatric microbes from diverse marine environments. Sci Data. 2018;5:180154.
Biller SJ, Berube PM, Dooley K, Williams M, Satinsky BM, Hackl T, et al. Marine microbial metagenomes sampled across space and time. Sci Data. 2018;5:180176.
Garczarek L, Guyet U, Doré H, Farrant GK, Hoebeke M, Brillet-Guéguen L, et al. Cyanorak v2.1: a scalable information system dedicated to the visualization and expert curation of marine and brackish picocyanobacteria genomes. Nucl Acids Res. 2021;49:D667–D676.
Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A, et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 2014;344:416–20.
Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N, et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA. 2013;110:9824–9.
Visintini N, Martiny AC, Flombaum P. Prochlorococcus, Synechococcus, and picoeukaryotic phytoplankton abundances in the global ocean. Limnol Oceanogr. 2021;6:207–15.
Paulsen ML, Doré H, Garczarek L, Seuthe L, Müller O, Sandaa R-A, et al. Synechococcus in the Atlantic gateway to the Arctic Ocean. Front Mar Sci. 2016;3:191.
Farrant GK, Doré H, Cornejo-Castillo FM, Partensky F, Ratin M, Ostrowski M, et al. Delineating ecologically significant taxonomic units from global patterns of marine picocyanobacteria. Proc Natl Acad Sci USA. 2016;113:E3365–74.
Sohm JA, Ahlgren NA, Thomson ZJ, Williams C, Moffett JW, Saito MA, et al. Co-occurring Synechococcus ecotypes occupy four major oceanic regimes defined by temperature, macronutrients and iron. ISME J. 2016;10:333–45.
Ahlgren NA, Rocap G. Diversity and distribution of marine Synechococcus: multiple gene phylogenies for consensus classification and development of qPCR assays for sensitive measurement of clades in the ocean. Front Microbiol. 2012;3:213–213.
Biller SJ, Berube PM, Lindell D, Chisholm SW. Prochlorococcus: The structure and function of collective diversity. Nat Rev Microbiol. 2015;13:13–27.
Huang S, Wilhelm SW, Harvey HR, Taylor K, Jiao N, Chen F. Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J. 2012;6:285–97.
Kent AG, Baer SE, Mouginot C, Huang JS, Larkin AA, Lomas MW, et al. Parallel phylogeography of Prochlorococcus and Synechococcus. ISME J. 2019;13:430–41.
Ustick LJ, Larkin AA, Garcia CA, Garcia NS, Brock ML, Lee JA, et al. Metagenomic analysis reveals global-scale patterns of ocean nutrient limitation. Science. 2021;372:287–91.
Martiny AC, Coleman ML, Chisholm SW. Phosphate acquisition genes in Prochlorococcus ecotypes: Evidence for genome-wide adaptation. Proc Natl Acad Sci USA. 2006;103:12552–7.
Martiny AC, Huang Y, Li W. Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ Microbiol. 2009;11:1340–7.
Kashtan N, Roggensack SE, Berta-Thompson JW, Grinberg M, Stepanauskas R, Chisholm SW. Fundamental differences in diversity and genomic population structure between Atlantic and Pacific Prochlorococcus. ISME J. 2017;11:1997–2011.
Zheng Y, Roberts RJ, Kasif S. Genomic functional annotation using co-evolution profiles of gene clusters. Genome Biol. 2002;3:research0060.1.
Overbeek R, Fonstein M, D’Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci USA. 1999;96:2896–901.
Kent AG, Dupont CL, Yooseph S, Martiny AC. Global biogeography of Prochlorococcus genome diversity in the surface ocean. ISME J. 2016;10:1856–65.
Song Q, Gordon AL, Visbeck M. Spreading of the Indonesian throughflow in the Indian Ocean. J Phys Oceanogr. 2004;34:772–92.
West NJ, Lebaron P, Strutton PG, Suzuki MT. A novel clade of Prochlorococcus found in high nutrient low chlorophyll waters in the South and Equatorial Pacific Ocean. ISME J. 2011;5:933–44.
Rusch DB, Martiny AC, Dupont CL, Halpern AL, Venter JC. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc Natl Acad Sci USA. 2010;107:16184–9.
Martiny AC, Kathuria S, Berube PM. Widespread metabolic potential for nitrite and nitrate assimilation among Prochlorococcus ecotypes. Proc Natl Acad Sci USA. 2009;106:10787–92.
Berube PM, Rasmussen A, Braakman R, Stepanauskas R, Chisholm SW. Emergence of trait variability through the lens of nitrogen assimilation in Prochlorococcus. eLife. 2019;8:e41043–43.
Berube PM, Biller SJ, Kent AG, Berta-Thompson JW, Roggensack SE, Roache-Johnson KH, et al. Physiology and evolution of nitrate acquisition in Prochlorococcus. ISME J. 2015;9:1195–207.
Burnat M, Li B, Kim SH, Michael AJ, Flores E. Homospermidine biosynthesis in the cyanobacterium Anabaena requires a deoxyhypusine synthase homologue and is essential for normal diazotrophic growth. Mol Microbiol. 2018;109:763–80.
Wang B, Xu Y, Wang X, Yuan JS, Johnson CH, Young JD, et al. A guanidine-degrading enzyme controls genomic stability of ethylene-producing cyanobacteria. Nat Commun. 2021;12:5150.
Nelson JW, Atilho RM, Sherlock ME, Stockbridge RB, Breaker RR. Metabolism of free guanidine in Bacteria is regulated by a widespread riboswitch class. Mol Cell. 2017;65:220–30.
Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, et al. RegPrecise 3.0 – a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genom. 2013;14:745.
Kamennaya NA, Post AF. Characterization of cyanate metabolism in marine Synechococcus and Prochlorococcus spp. Appl Environ Microbiol. 2011;77:291–301.
Kamennaya NA, Post AF. Distribution and expression of the cyanate acquisition potential among cyanobacterial populations in oligotrophic marine waters. Limnol Oceanogr. 2013;58:1959–71.
Jones LB, Ghosh P, Lee J-H, Chou C-N, Kunz DAY. Linkage of the Nit1C gene cluster to bacterial cyanide assimilation as a nitrogen source. Microbiol. 2018;164:956–68.
Pablo F, Stauber JL, Buckney RT. Toxicity of cyanide and cyanide complexes to the marine diatom Nitzschia closterium. Water Res. 1997;31:2435–42.
Saunders JK, Rocap G. Genomic potential for arsenic efflux and methylation varies among global Prochlorococcus populations. ISME J. 2016;10:197–209.
Riedel GF. Influence of salinity and sulfate on the toxicity of chromium(vi) to the estuarine diatom Thalassiosira pseudonana. J Phycol. 1984;20:496–500.
Pernil R, Picossi S, Mariscal V, Herrero A, Flores E. ABC-type amino acid uptake transporters Bgt and N-II of Anabaena sp. strain PCC 7120 share an ATPase subunit and are expressed in vegetative cells and heterocysts. Mol Microbiol. 2008;67:1067–80.
Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF, et al. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–70.
Moore CM, Mills MM, Arrigo KR, Berman-Frank I, Bopp L, Boyd PW, et al. Processes and patterns of oceanic nutrient limitation. Nat Geosci. 2013;6:701–10.
Tetu SG, Brahamsha B, Johnson DA, Tai V, Phillippy K, Palenik B, et al. Microarray analysis of phosphate regulation in the marine cyanobacterium Synechococcus sp. WH8102. ISME J. 2009;3:835–49.
Clark LL, Ingall ED, Benner R. Marine phosphorus is selectively remineralized. Nature. 1998;393:426–426.
Feingersch R, Philosof A, Mejuch T, Glaser F, Alalouf O, Shoham Y, et al. Potential for phosphite and phosphonate utilization by Prochlorococcus. ISME J. 2012;6:827–34.
Martinez A, Tyson GW, Delong EF. Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ Microbiol. 2010;12:222–38.
Sosa OA, Casey JR, Karl DM. Methylphosphonate oxidation in Prochlorococcus strain MIT9301 supports phosphate acquisition, formate excretion, and carbon assimilation into purines. Appl Environ Microbiol. 2019;85:e00289–19.
Martínez A, Osburne MS, Sharma AK, DeLong EF, Chisholm SW. Phosphite utilization by the marine picocyanobacterium Prochlorococcus MIT9301. Environ Microbiol. 2012;14:1363–77.
McSorley FR, Wyatt PB, Martinez A, DeLong EF, Hove-Jensen B, Zechel DL. PhnY and PhnZ comprise a new oxidative pathway for enzymatic cleavage of a carbon–phosphorus bond. J Am Chem Soc. 2012;134:8364–7.
Acker M, Hogle SL, Berube PM, Hackl T, Coe A, Stepanauskas R, et al. Phosphonate production by marine microbes: exploring new sources and potential function. Proc Natl Acad Sci USA. 2022;119:e2113386119.
Malmstrom RR, Rodrigue S, Huang KH, Kelly L, Kern SE, Thompson A, et al. Ecology of uncultured Prochlorococcus clades revealed through single-cell genomics and biogeographic analysis. ISME J. 2013;7:184–98.
Cooley JW, Vermaas WFJ. Succinate dehydrogenase and other respiratory pathways in thylakoid membranes of Synechocystis sp. strain PCC 6803: capacity comparisons and physiological function. J Bacteriol. 2001;183:4251–8.
Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68.
Buffet A, Rocha EPC, Rendueles O. Nutrient conditions are primary drivers of bacterial capsule maintenance in Klebsiella. Proc R Soc B Biol Sci. 2021;288:20202876.
Lea-Smith DJ, Ross N, Zori M, Bendall DS, Dennis JS, Scott SA, et al. Thylakoid terminal oxidases are essential for the cyanobacterium Synechocystis sp. PCC 6803 to survive rapidly changing light intensities. Plant Physiol. 2013;162:484–95.
Lea-Smith DJ, Bombelli P, Vasudevan R, Howe CJ. Photosynthetic, respiratory and extracellular electron transport pathways in cyanobacteria. Biochim Biophys Acta Bioenerg. 2016;1857:247–55.
Kranzler C, Lis H, Finkel OM, Schmetterer G, Shaked Y, Keren N. Coordinated transporter activity shapes high-affinity iron acquisition in cyanobacteria. ISME J. 2014;8:409–17.
Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, et al. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. 2009;73:249–99.
Ford BA, Ranjit P, Mabbutt BC, Paulsen IT, Shah BS. ProX from marine Synechococcus spp. show a sole preference for glycine-betaine with differential affinity between ecotypes. Environ Microbiol. 2022;24:6071–85.
Dempwolff F, Wischhusen HM, Specht M, Graumann PL. The deletion of bacterial dynamin and flotillin genes results in pleiotrophic effects on cell division, cell growth and in cell shape maintenance. BMC Microbiol. 2012;12:298.
Boulay C, Wilson A, D’Haene S, Kirilovsky D. Identification of a protein required for recovery of full antenna capacity in OCP-related photoprotective mechanism in cyanobacteria. Proc Natl Acad Sci USA. 2010;107:11620–5.
Six C, Ratin M, Marie D, Corre E. Marine Synechococcus picocyanobacteria: light utilization across latitudes. Proc Natl Acad Sci USA. 2021;118:e2111300118.
Xia X, Partensky F, Garczarek L, Suzuki K, Guo C, Yan Cheung S, et al. Phylogeography and pigment type diversity of Synechococcus cyanobacteria in surface waters of the northwestern Pacific Ocean. Environ Microbiol. 2017;19:142–58.
Grébert T, Doré H, Partensky F, Farrant GK, Boss ES, Picheral M, et al. Light color acclimation is a key process in the global ocean distribution of Synechococcus cyanobacteria. Proc Natl Acad Sci USA. 2018;115:E2010–19.
Sanfilippo JE, Nguyen AA, Karty JA, Shukla A, Schluchter WM, Garczarek L, et al. Self-regulating genomic island encoding tandem regulators confers chromatic acclimation to marine Synechococcus. Proc Natl Acad Sci USA. 2016;113:6077–82.
Larkin AA, Martiny AC. Microdiversity shapes the traits, niche space, and biogeography of microbial taxa: the ecological function of microdiversity. Environ Microbiol Rep. 2017;9:55–70.
Barton NH. Genetic hitchhiking. Philos Trans R Soc Lond Ser B: Biol Sci. 2000;355:1553–62.
Wiedenbeck J, Cohan FM. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev. 2011;35:957–76.
Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, et al. Structure and function of the global ocean microbiome. Science. 2015;348:1261359–59.
Szmrecsanyi B. Grammatical variation in British English dialects: a study in corpus-based dialectometry. 2012. Cambridge University Press, Cambridge.
Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinfo. 2008;9:559.
Csardi G, Nepusz T. The igraph software package for complex network research. InterJ, Complex Syst. 2006;1695:1–9.
Acknowledgements
This work was supported by the French “Agence Nationale de la Recherche” Programs SAMOSA (ANR-13-ADAP-0010), CINNAMON (ANR‐17‐CE02‐0014), EFFICACY (ANR-19-CE02-0019), and France Génomique (ANR-10-INBS-09) as well as the European Union program Assemble + (Horizon 2020, under grant agreement number 287589). We acknowledge Christophe Six for his help with cloning some of the Synechococcus strains used in this study and Francisco M. Cornejo-Castillo for helpful discussions. We also thank the support and commitment of the Tara Oceans coordinators and consortium, Agnès b. and E. Bourgois, the Veolia Environment Foundation, Région Bretagne, Lorient Agglomeration, World Courier, Illumina, the EDF Foundation, FRB, the Prince Albert II de Monaco Foundation, the Tara schooner, and its captains and crew. Tara Oceans would not exist without continuous support from 23 institutes (http://oceans.taraexpeditions.org).
Author information
Authors and Affiliations
Contributions
HD and LG conceived and supervised the project. MR, MO, GKF, DJS, FP, LG, and PW participated in the generation of genomes and/or metagenomes used in this study. UG, HD, GKF, and JL performed the bioinformatics analyses. LBG, MH, JS, GL, and EC developed the Cyanorak information system and FP, HD, UG, JL, MF, MO, DJS, and LG curated the genome database. UG, BA, and DE performed the network and statistical analyses. HD, UG, FP, DJS, DE, and LG wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Doré, H., Guyet, U., Leconte, J. et al. Differential global distribution of marine picocyanobacteria gene clusters reveals distinct niche-related adaptive strategies. ISME J 17, 720–732 (2023). https://doi.org/10.1038/s41396-023-01386-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-023-01386-0