Abstract
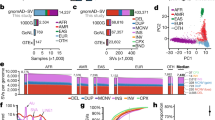
Establishing the age of each mutation segregating in contemporary human populations is important to fully understand our evolutionary history1,2 and will help to facilitate the development of new approaches for disease-gene discovery3. Large-scale surveys of human genetic variation have reported signatures of recent explosive population growth4,5,6, notable for an excess of rare genetic variants, suggesting that many mutations arose recently. To more quantitatively assess the distribution of mutation ages, we resequenced 15,336 genes in 6,515 individuals of European American and African American ancestry and inferred the age of 1,146,401 autosomal single nucleotide variants (SNVs). We estimate that approximately 73% of all protein-coding SNVs and approximately 86% of SNVs predicted to be deleterious arose in the past 5,000–10,000 years. The average age of deleterious SNVs varied significantly across molecular pathways, and disease genes contained a significantly higher proportion of recently arisen deleterious SNVs than other genes. Furthermore, European Americans had an excess of deleterious variants in essential and Mendelian disease genes compared to African Americans, consistent with weaker purifying selection due to the Out-of-Africa dispersal. Our results better delimit the historical details of human protein-coding variation, show the profound effect of recent human history on the burden of deleterious SNVs segregating in contemporary populations, and provide important practical information that can be used to prioritize variants in disease-gene discovery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Data deposits
Filtered sets of annotated variants and their allele frequencies are available at (http://evs.gs.washington.edu/EVS/) and genotypes and phenotypes from a large subset of individuals are also available through dbGaP (http://www.ncbi.nlm.nih.gov/gap) using the following accession information: NHLBI GO-ESP: Women’s Health Initiative Exome Sequencing Project (WHI) – WHISP, WHISP_Subject_Phenotypes, pht002246.v2.p2, phs000281.v2.p2; NHLBI GO-ESP: Heart Cohorts Exome Sequencing Project (JHS), ESP_HeartGO_JHS_LDLandEOMI_Subject_Phenotypes, pht002539.v1.p1, phs000402.v1.p1; NHLBI GO-ESP: Heart Cohorts Exome Sequencing Project (FHS), HeartGO_FHS_LDLandEOMI_PhenotypeDataFile, pht002476.v1.p1, phs000401.v1.p1; NHLBI GO-ESP: Heart Cohorts Exome Sequencing Project (CHS), HeartGO_CHS_LDL_PhenotypeDataFile, pht002536.v1.p1, phs000400.v1.p1; NHLBI GO-ESP: Heart Cohorts Exome Sequencing Project (ARIC), ESP_ARIC_LDLandEOMI_Sample, pht002466.v1.p1, phs000398.v1.p1;NHLBIGO-ESP: Lung Cohorts Exome Sequencing Project (Cystic Fibrosis), ESP_LungGO_CF_PA_Culture_Data, pht002227.v1.p1, phs000254.v1.p1; NHLBI GO-ESP: Early-Onset Myocardial Infarction (Broad EOMI), ESP_Broad_EOMI_Subject_Phenotypes, pht001437.v1.p1, phs000279.v1.p1; NHLBI GO-ESP: Lung Cohorts Exome Sequencing Project (Pulmonary Arterial Hypertension), PAH_Subject_Phenotypes_Baseline_Measures, pht002277.v1.p1, phs000290.v1.p1; NHLBI GO-ESP: Lung Cohorts Exome Sequencing Project (Lung Health Study of Chronic Obstructive Pulmonary Disease), LHS_COPD_Subject_Phenotypes_Baseline_Measures, pht002272.v1.p1, phs000291.v1.p1.
References
Kimura, M. & Ota, T. The age of a neutral mutant persisting in a finite population. Genetics 75, 199–212 (1973)
Tishkoff, S. A. & Verrelli, B. C. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu. Rev. Genomics Hum. Genet. 4, 293–340 (2003)
Slatkin, M. & Rannala, B. Estimating allele age. Annu. Rev. Genomics Hum. Genet. 1, 225–249 (2000)
Keinan, A. & Clark, A. G. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science 336, 740–743 (2012)
Nelson, M. R. et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 337, 100–104 (2012)
Tennessen, J. A. et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337, 64–69 (2012)
Griffiths, R. C. & Tavaré, S. The age of a mutation in a general coalescent tree. Commun. Stat. Stoch. Models 14, 273–295 (1998)
Coventry, A. et al. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nature Commun. 1, 131 (2010)
Gravel, S. et al. Demographic history and rare allele sharing among human populations. Proc. Natl Acad. Sci. USA 108, 11983–11988 (2011)
Gutenkunst, R. N., Hernandez, R. D., Williamson, S. H. & Bustamante, C. D. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5, e1000695 (2009)
Schaffner, S. F. et al. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 15, 1576–1583 (2005)
Gibson, G. Rare and common variants: twenty arguments. Nature Rev. Genet. 13, 135–145 (2012)
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols 4, 1073–1081 (2009)
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nature Methods 7, 248–249 (2010)
Chun, S. & Fay, J. C. Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561 (2009)
Schwarz, J. M., Rodelsperger, C., Schuelke, M. & Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nature Methods 7, 575–576 (2010)
Davydov, E. V. et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLOS Comput. Biol. 6, e1001025 (2010)
Pollard, K. S., Hubisz, M. J., Rosenbloom, K. R. & Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010)
Becker, K. G., Barnes, K. C., Bright, T. J. & Wang, S. A. The genetic association database. Nature Genet. 36, 431–432 (2004)
Pyun, J. A., Cha, D. H. & Kwack, K. LAMC1 gene is associated with premature ovarian failure. Maturitas 71, 402–406 (2012)
Liu, Q. et al. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron 56, 66–78 (2007)
Jia, E. Z. et al. Association of the mutation for the human carboxypeptidase E gene exon 4 with the severity of coronary artery atherosclerosis. Mol. Biol. Rep. 36, 245–254 (2009)
Valdmanis, P. N. et al. Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am. J. Hum. Genet. 80, 152–161 (2007)
Blekhman, R. et al. Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 18, 883–889 (2008)
Liao, B. Y., Scott, N. M. & Zhang, J. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol. Biol. Evol. 23, 2072–2080 (2006)
Lohmueller, K. E. et al. Proportionally more deleterious genetic variation in European than in African populations. Nature 451, 994–997 (2008)
Hawks, J., Wang, E. T., Cochran, G. M., Harpending, H. C. & Moyzis, R. K. Recent acceleration of human adaptive evolution. Proc. Natl Acad. Sci. USA 104, 20753–20758 (2007)
Acknowledgements
We acknowledge the support of the National Heart, Lung and Blood Institute (NHLBI), the contributions of the research institutions that participated in this study, the study investigators, field staff and study participants who created this resource for biomedical research, and the Population Genetics Project Team of the NHLBI. We thank J. Wilson and R. Do for critical feedback on the manuscript. Funding for the GO (Grand Opportunity) Exome Sequencing Project was provided by NHLBI grants RC2 HL-103010 (Heart GO), RC2 HL-102923 (Lung GO) and RC2 HL-102924 (WHISP). The exome sequencing was was supported by NHLBI grants RC2 HL-102925 (Broad GO) and RC2 HL-102926 (Seattle GO).
Author information
Authors and Affiliations
Contributions
W.F. and J.M.A. conceived the analyses. D.A.N., S.G., M.J.R. and D.A. oversaw data generation and quality control. G.J., H.M.K. and G.A. developed algorithms and identified SNVs from the sequencing data. W.F. carried out the majority of analyses with contributions from T.D.O. W.F., M.J.B., J.S. and J.M.A. analysed the data and wrote the manuscript with contributions from all authors. W.F., T.D.O., S.M.L., J.S., M.J.R., D.A.N., M.J.B. and J.M.A. are members of the Seattle Grand Opportunity (GO) group and G.J., H.M.K., G.A., S.G. and D.A. are members of the Broad GO group, which are both sub-groups of the NHLBI Exome Sequencing Project (ESP).
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Text and Data, Supplementary References, Supplementary Tables 1-4 and Supplementary Figures 1-15 (see Table of Contents for more details). (PDF 3066 kb)
Rights and permissions
About this article
Cite this article
Fu, W., O’Connor, T., Jun, G. et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493, 216–220 (2013). https://doi.org/10.1038/nature11690
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11690
This article is cited by
-
Novel MTR compound-heterozygous mutations in a Chinese girl with HHcy due to methionine synthase deficiency, cblG: a case report
Egyptian Journal of Medical Human Genetics (2024)
-
Pathogenic variants in human DNA damage repair genes mostly arose in recent human history
BMC Cancer (2024)
-
Evolutionary origin of germline pathogenic variants in human DNA mismatch repair genes
Human Genomics (2024)
-
ClinVar and HGMD genomic variant classification accuracy has improved over time, as measured by implied disease burden
Genome Medicine (2023)
-
Lack of CFAP54 causes primary ciliary dyskinesia in a mouse model and human patients
Frontiers of Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.