Abstract
Bread wheat (Triticum aestivum) is a globally important crop, accounting for 20 per cent of the calories consumed by humans. Major efforts are underway worldwide to increase wheat production by extending genetic diversity and analysing key traits, and genomic resources can accelerate progress. But so far the very large size and polyploid complexity of the bread wheat genome have been substantial barriers to genome analysis. Here we report the sequencing of its large, 17-gigabase-pair, hexaploid genome using 454 pyrosequencing, and comparison of this with the sequences of diploid ancestral and progenitor genomes. We identified between 94,000 and 96,000 genes, and assigned two-thirds to the three component genomes (A, B and D) of hexaploid wheat. High-resolution synteny maps identified many small disruptions to conserved gene order. We show that the hexaploid genome is highly dynamic, with significant loss of gene family members on polyploidization and domestication, and an abundance of gene fragments. Several classes of genes involved in energy harvesting, metabolism and growth are among expanded gene families that could be associated with crop productivity. Our analyses, coupled with the identification of extensive genetic variation, provide a resource for accelerating gene discovery and improving this major crop.
Similar content being viewed by others
Main
With a global output of 681 million tonnes in 20111, bread wheat accounts for 20% of the calories consumed by humans2 and is an important source of protein, vitamins and minerals. It originated from hybridization between cultivated tetraploid emmer wheat (AABB, Triticum dicoccoides) and diploid goat grass (DD, Aegilops tauschii) approximately 8,000 years ago3. Bread wheat cultivation and domestication has been directly associated with the spread of agriculture and settled societies, and it is now one of the most widely cultivated crops owing to its high yields and nutritional and processing qualities. The three diploid progenitor genomes, AA from Triticum urartu, BB from a species that is unknown but which may be of the section Sitopsis (to which Aegilops speltoides belongs), and DD from Ae. tauschii, radiated from a common Triticeae ancestor between 2.5 and 4.5 million years ago, and AABB tetraploids arose less than 0.5 million years ago4,5. Nucleotide diversity in the AABB and DD genomes is substantially reduced compared with ancestral populations, indicating a major diversity bottleneck on the transition to cultivated lines6.
Grass genomes show extensive long-range conservation of gene order7,8,9. Nevertheless, they are highly dynamic owing to the activities of repeats that contribute to tremendous variation in genome size10, changes in local gene order and pseudogene formation, particularly in larger genomes such as those of maize11 and wheat12. From analysis of BAC contigs on chromosome 3B, the 17-gigabase-pair (Gb) genome was estimated to be composed of approximately 80% repeats, primarily retroelements, with a gene density of between 1 per 87 kilobase pairs and 1 per 184 kilobase pairs13. Despite both the substantial knowledge gained of the wheat genome from these studies and the central importance of the wheat crop, a comprehensive genome-wide analysis of gene content has yet to be conducted owing to its large size, repeat content and polyploid complexity.
We have analysed a low-coverage, long-read (454) shotgun sequence of the hexaploid wheat genome using gene sequences from diverse grasses. From this, we created assemblies of wheat genes in an orthologous gene family framework, used diploid wheat relatives to classify homeologous relationships, and defined a genome-wide catalogue of single nucleotide polymorphisms (SNPs) in the A, B and D genomes. These analyses provide a foundation for genetic and genomic analysis of this key crop.
Sequence analysis
The wheat variety Chinese Spring (CS42) was selected for sequencing because of its wide use in genome studies14,15. Purified nuclear DNA was sequenced using Roche 454 pyrosequencing technology (GS FLX Titanium and GS FLX+ platforms) to generate 85 Gb of sequence (220 million reads), corresponding to approximately fivefold coverage on the basis of an estimated genome size of 17 Gb. Supplementary Table 1 shows that 79% of the reads had matches to the Triticeae Repeat Sequence Database, and most hit retrotransposons, consistent with previous studies13. To identify A-, B- and D-genome-derived gene assemblies in the hexaploid sequences, we used Illumina sequence assemblies of Triticum monococcum, related to the A-genome donor, Ae. speltoides complementary DNA (cDNA) assemblies and 454 sequences from the D-genome donor Ae. tauschii, respectively. The SOLiD platform was used to generate additional sequence of CS42 and three commercial wheat varieties to increase the accuracy of homeologous SNP identification. Data sets are summarized in Table 1 and Supplementary Table 2, and SNP identification methods are described in Supplementary Information, section 5.2.
Sequence assembly
An orthologous group assembly (Supplementary Table 3) was created by clustering 454 reads by sequence similarity to orthologous grass gene sequences, and separate assembly of the clusters at high stringency using Newbler (Supplementary Information, section 2). The orthologous genes were derived from rice16, sorghum8, Brachypodium9 and barley full-length cDNAs by OrthoMCL17 clustering. This generated 20,496 orthologous groups (Supplementary Table 4 and Supplementary Fig. 1). The gene model with highest similarity to wheat (termed the orthologous group representative (OGR)) was selected from each orthologous group by stringent BLASTX comparison to a low-copy-number genome assembly (LCG) made by filtering out repetitive sequences and assembling the remaining low-copy-number sequences de novo (Supplementary Table 3). The assemblies are described in Table 2. Nearly 90% of the metabolic genes in Arabidopsis matched OGRs, and the 20,051 OGRs matched 92% of publicly available wheat full-length cDNAs18 and 78.7% of the harvEST set of wheat cDNA assemblies (Supplementary Fig. 2), indicating that they represent nearly all wheat genes.
We optimized parameters for wheat gene assembly using MetaSim19 to generate simulated fivefold 454 reads from the allotetraploid maize genome and from a triplicated rice gene set, with the introduction of sequence variation (Supplementary Information, section 2.7). Similar degrees of coverage over the OGRs were seen for the simulated data sets and wheat 454 reads (Fig. 1a). Rice reads followed the same depth distribution as the wheat reads (Fig. 1b), suggesting that they are a reasonable representation of hexaploid sequences. Maize reads covered their OGRs to a median depth of approximately five, consistent with fivefold coverage.
a, Coverage of OGRs by repeat-masked wheat 454 sequence reads (black line), wheat LCG (black dashed line) and the orthologous group assembly (blue line), together with rice genes (red line) and maize simulated reads (green line). b, Median coverage depth over protein-coding regions of OGRs (amino terminus = 0; carboxy terminus = 100). The colour coding is the same as in a, except simulated hexaploid reads from rice (red line) were used. c, Distribution of wheat gene copy numbers from the orthologous group assembly.
Simulated maize and triplicated rice 454 reads were used to optimize assembly parameters. Assembly at 99% minimum sequence identity (m.i.) using 40-bp overlap length predicted gene family sizes most accurately (Supplementary Figs 3–6). Wheat 454 reads were preprocessed (Supplementary Table 5) and assembled using 99% m.i. (Supplementary Tables 6 and 7) to create the orthologous group assembly. Figure 1b shows that the depth of coverage of the orthologous group assembly followed a similar pattern to maize, consistent with multiple gene copies. In contrast, the low depth coverage by the LCG assembly suggested that gene family numbers were collapsed. The number of wheat assemblies for each OGR was calculated to determine gene copy numbers (Supplementary Table 7). Figure 1c shows that most OGRs had between one and five distinctive wheat gene assemblies, with a peak of two genes.
The A, B and Ae. tauschii (D) genomes13,20,21 have been estimated to contain approximately 28,000, 38,000 and 36,000 genes, respectively. We estimated the number of genes in the hexaploid wheat genome to range between 94,000 and 96,000 (Supplementary Information, section 2.10). This is reasonably consistent with estimates based on wheat chromosome sequences13. Comparing our transcriptome assembly (Supplementary Information, sections 2.8 and 2.9) and wheat harvEST with the wheat OGRs showed that 76% and, respectively, 65% were expressed under the conditions used for RNA isolation. Similar results were found in barley22, rice16 and maize23, indicating that the assemblies are bona fide wheat genes.
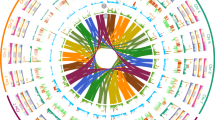
We defined the overall extent of gene conservation between wheat and the most closely related sequenced pooid grass, Brachypodium distachyon9,24. Track 1 of Fig. 2 shows that there is a high degree of overlap between the gene sets of Brachypodium and wheat, but with regions of lower conservation, for example on Brachypodium chromosomes 1 and 4. Syntenic maps of the Brachypodium genome and the A-, B- and D-chromosome groups were created by integrating high-density wheat EST-based markers25 with Brachypodium genes (Fig. 2, tracks 5, 6 and 7, respectively). Supplementary Fig. 7 shows the A-, B- and D-genome markers separately. Syntenic alignments were readily identifiable and conformed to the predicted major patterns9,26. We identified many insertions and/or translocations of blocks of genes within the overall conserved patterns of gene order, including the major rearrangement on chromosome 4A as shown on Brachypodium chromosome 1 (ref. 20). Lower marker density on the D genome is evident in track 7. The higher-resolution genetic map identified a new syntenic alignment of Triticeae group 5 to Brachypodium chromosome 3 genes.
The inner circle represent gene order on the five Brachypodium chromosomes (Bd1–Bd5). Track 1 illustrates conservation between wheat 454 reads and Brachypodium genes, shown as a window of genes present in wheat. Tracks 2–4 show SNP density (the mean number of SNPs per gene in a window of 20 genes) in the A (track 2), B (track 3) and D (track 4) genomes of wheat. Tracks 5–7 show wheat synteny with Brachypodium for the A (track 5), B (track 6) and D (track 7) genomes. Genetic markers25 (shown in darker colours) are colour-coded by wheat chromosome. Gaps between markers are filled in to show synteny (lighter colours).
Genome change in polyploid wheat
We determined the influence of polyploidy on gene content in hexaploid wheat by defining the sizes of gene families in hexaploid wheat and the diploid progenitor Ae. tauschii from the copy number of genes for each OGR, which were then paired with the gene family size of the OGR in sequenced diploid grasses (Supplementary Information, section 2.6). The mean family size was 1.4 members. Supplementary Fig. 8 shows relationships between wheat and diploid orthologous gene family across the full scale of orthologous gene family sizes. This approach accurately reconstructed gene family sizes in simulated maize and ‘hexaploid’ rice genomes (Figs 3a, b), although larger gene family sizes tended to be underestimated. Figure 3c, d shows the relationships between Ae. tauschii and wheat genes. Single-member gene families in hexaploid wheat and Ae. tauschii were maintained to a similar extent as those seen in sequenced diploid grasses, consistent with Southern blot analyses of single-copy genes27. Using the D genome as a diploid reference, we calculated the Triticeae hexaploid/diploid gene family size ratio to be between 2.5:1 and 2.7:1, derived from the geometric mean (2.5:1) and the slopes of the blue line and the red line (2.7:1) in Fig. 3e. Comparing this with the expected hexaploid/diploid ratio of 3:1 indicates the loss of between 10,000 and 16,000 genes in hexaploid wheat compared with the three diploid progenitors (Supplementary Information, section 2.10). This is consistent with earlier studies of gene loss in newly synthesized wheat polyploids28 and the erosion of genetic diversity during wheat domestication6.
The boxes and whiskers contain 50% and 90% of the orthologous group assembly genes, respectively. The box colours indicate the number of genes in diploid gene families of different sizes. The black lines represent expected gene family sizes, and the red lines show the gene family sizes determined from the orthologous group assembly, derived by polynomial regression fit. Only gene families with up to ten members are shown. a, Maize gene family sizes predicted from orthologous assembly of simulated 454 reads. b, Rice gene family sizes predicted from orthologous assembly of simulated 454 reads derived from triplicated rice genes. c, Aegilops tauschii gene family sizes obtained from orthologous assembly of repeat-masked 454 reads. Expanded gene families are shown as green dots. d, Wheat gene family sizes in the orthologous group assembly. e, Amalgamation of wheat and Ae. tauschii gene copy numbers. The black line shows the respective expected gene copy numbers for wheat and Ae. tauschii. The red line shows the regression fit for wheat, and the blue line shows the regression fit for Ae. tauschii. The grey zone between these lines estimates the extent of gene loss in hexaploid wheat. For each family size, the left-hand boxes represent hexaploid wheat and the right-hand boxes represent Ae. tauschii.
Despite this overall trend of gene family size reduction, gene families with fewer or more members than expected were identified in Ae. tauschii and hexaploid wheat, as shown by green dots (more members) and brown dots (fewer members) in Fig. 3c (Ae. tauschii) and Fig. 3d (hexaploid wheat). Supplementary Tables 10–12 show the over- and under-represented functional categories of protein. Most of the over-represented categories in expanded gene families are common to wheat and Ae. tauschii: these include ribosome proteins, components of photosystem II, storage proteins, transposon-related proteins, cytochrome P450s, NB-ARC domain proteins involved in defence responses, proteins related to pollen allergens and F-box proteins. Five of the eleven families encoding hydrogen ion transmembrane transporters were significantly more numerous in Ae. tauschii than in wheat. Analysis of gene families (Supplementary Fig. 9) showed that they encode different subunits of ATPases. We speculate that they may provide proton gradients to support Na+ exclusion in Ae. tauschii29 and the accumulation of minerals in other Aegilops species30.
Pseudogene analysis
Several classes of plant DNA transposons31,32 and retroelements33 create and amplify gene fragments, disrupt genes and create pseudogenes, which can influence gene expression through epigenetic mechanisms34. We identified a set of almost 233,000 gene fragments that mapped to the same regions of their OGRs, forming ‘stacks’ that were sufficiently divergent not to assemble into their cognate gene assemblies (Fig. 4a). Two classes were identified: those containing Pfam domains and those aligning with non-Pfam domains of OGRs. Nearly 30% of the OGRs had associated gene fragments (Supplementary Table 13) that most frequently covered between 5 and 15% of the OGR length (Fig. 4b). Figure 4c shows that the alignment identities of gene fragments against their OGRs were substantially lower than the identities of cognate regions within wheat gene assemblies. Supplementary Fig. 10 shows the distribution of stacks along genes and the ratio of non-synonymous to synonymous substitutions (Ka/Ks) along the genes. Pfam domains found in stacks were enriched for zinc-finger motifs in mutator transposons (Supplementary Table 14), consistent with their role in pseudogene formation31. F-box, protein kinase and NB-ARC domains, which are found in the most rapidly evolving gene families in plants9,35, are also over-represented.
a, Visualization of an OGR and associated wheat sequences. The top track shows the hit count profile of mapped 454 reads. The lower tracks show subassemblies of three wheat genes and a stacked region of gene fragments. Read depth is represented by the heat map. b, Coverage of the OGR by Pfam-containing gene fragments and pseudogenes. The blue and red lines represent stacks with and without protein domains, respectively. c. Protein identity between subassemblies forming stacks of gene fragments. The blue and red lines represent stacks with and without protein domains, respectively, and the black line represents subassemblies forming genes.
Determining homeologous relationships of gene assemblies
We classified gene assemblies as A-, B- or D-genome-derived according to sequence similarity to Illumina sequence assemblies from T. monococcum, cDNA assemblies from Ae. speltoides and, respectively, 454 reads from Ae. tauschii by applying a support vector machine learning approach (Supplementary Section 5, Supplementary Figs 11 and 12, and Supplementary Tables 15–18). Supplementary Fig. 13 shows that 66% of the gene assemblies were classified with high overall precision (>70%) and recall into the A genome (28.3%), the B genome (29.2%) and the D genome (33.8%). The other 9% of classified assemblies have stop codons. The othes 34% with low classification probabilities are likely to be very similar homeologues. Comparison with a subset of A-, B- and D-genome SNPs confirmed 72% of A-genome classifications and 85% of D-genome classifications (Fig. 2 and Supplementary Table 19). Discrimination of putative B-genome genes was only ∼60%, possibly owing both to the use of cDNA sequences for classification when most of the informative sequence polymorphisms are intronic, and to uncertainty about the ancestry of the B genome5. The set of 132,552 SNPs allocated to the A, B and D genomes is displayed using Brachypodium as a template in tracks 2–4 of Fig. 2.
There were no significant differences between the respective distributions of GO Slim molecular function categories in the A, B and D genes (Supplementary Fig. 14), indicating that at this level of functional categorization there is no biased gene loss36 in any of the genomes. Nevertheless, analysis of GO Slim terms associated with stop codons in A, B and D gene assemblies showed that there was a strong tendency to retain functional copies of genes encoding transcription factors in all three genomes (Supplementary Fig. 15), similar to the preferential retention of these genes in Arabidopsis genome duplications37. This indicates that genome-specific transcriptional regulatory networks tend to be maintained in wheat.
Conclusions
Using whole-genome 454 sequencing, we assembled gene sequences representing an essentially complete gene set, and a significant number were assigned to the A, B or D genome. Although the assemblies are fragmentary, they form a powerful framework for identifying genes, accelerating further genome sequencing and facilitating genome-scale analyses. The identification of over 132,000 SNPs in A, B and D genes facilitates analysis of quantitative trait loci and association studies of traits. Comparison with the sequences of diploid progenitors and relatives showed pronounced reductions in the size of large gene families in wheat despite the relatively recent formation of the hexaploid (Fig. 3e), consistent with smaller-scale analyses28,38. The scale of gene loss in hexaploid wheat compared with maize36 and Brassica rapa39 is significantly smaller, possibly as a result of its relatively recent origin and the absence of intergenome recombination40. Nevertheless, gene loss in wheat could be rapid, as shown in the newly created allopolyploid Tragopogon miscellus41. Most functional classes show equal gene loss in the three genomes, but families of transcription factors showed a clear tendency to be retained as functional genes in all three genomes. These may maintain transcriptional networks in each genome and contribute to non-additive gene expression42 and genome plasticity. In contrast to the overall loss of gene family members, several classes of gene families with predicted roles in defence, nutritional content, energy metabolism and growth have increased sizes in the Triticeae lineage, possibly as a result of selection during domestication.
Major efforts are underway to improve wheat productivity by increasing genetic diversity in breeding materials and through genetic analysis of traits43. The genomic resources that we have developed promise to accelerate progress by facilitating the identification of useful variation in genes of wheat landraces and progenitor species, and by providing genomic landmarks to guide progeny selection. Analysis of complex polygenic traits such as yield and nutrient use efficiency will also be accelerated, contributing to sustainable increases in wheat crop production.
Methods Summary
A single-seed descent line of T. aestivum landrace Chinese Spring was sequenced, because it is widely used for cytogenetic analysis44 and physical mapping15. Triticum monococcum accession 4342-96 is a community standard line for targeting induced local lesions in genomes, physical mapping and genetic analysis; and Ae. tauschii ssp strangulata accession AL8/78, which is used for physical and genetic mapping, was sequenced using 454 technology.
Sequence for the T. aestivum wheat gene assembly was generated using Roche 454 pyrosequencing on the GS FLX Titanium and GS FLX+ platforms. Additional sequence read data sets for T. aestivum, T. monococcum and Ae. tauschii were generated using three platforms, Illumina, 454 and SOLiD, to analyse homeologous sequences and SNPs (a list of all data sets is in Supplementary Table 2). Orthologous groups were created from rice, sorghum and B. distachyon genome sequences and barley full-length cDNA sequences. Wheat gene assemblies were named according to their OGR and were identified by a seven-digit identifier and their predicted genome (for example Traes_Bradi1g12345_0000001_D and Traes_Sb3g33333_6543210_A). Gene and cDNA assemblies can be searched at the MIPS Wheat Genome Database (http://mips.helmholtz-muenchen.de/plant/wheat/uk454survey/index.jsp). All sequence data has been deposited in publicly accessible databases, described in Supplementary Information. Sequence assemblies, annotated gene sequences and their relationships are available for download from the European Bioinformatics Institute (http://www.ebi.ac.uk) and viewing in a synteny-based Ensembl genome browser. Annotated gene sequences and their relationships can be viewed in a Brachypodium synteny-based Ensembl genome browser (http://plants.ensembl.org/brachypodium_distachyon).
References
United States. Department of Agriculture. World Agricultural Supply and Demand Estimates. Report No. WASDE-511; http://usda01.library.cornell.edu/usda/current/wasde/wasde-10-11-2012.pdf (2012)
Food and Agriculture Organisation of the United Nations. http://faostat.fao.org/default.aspx?lang=en (2011)
Nesbitt, M. & Samuel, D. in Hulled Wheats (eds Padulosi, S., Hammer, K. & Heller, J. ) 41–100 (Proc. 1st Internat. Workshop Hulled Wheats, International Plant Genetic Resources Institute, 1996)
Dvorak, J., Akhunov, E. D., Akhunov, A. R., Deal, K. R. & Luo, M. C. Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol. Biol. Evol. 23, 1386–1396 (2006)
Salse, J. et al. New insights into the origin of the B genome of hexaploid wheat: evolutionary relationships at the SPA genomic region with the S genome of the diploid relative Aegilops speltoides. BMC Genomics 9, 555 (2008)
Haudry, A. et al. Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol. Biol. Evol. 24, 1506–1517 (2007)
Moore, G., Devos, K. M., Wang, Z. & Gale, M. D. Cereal genome evolution. Grasses, line up and form a circle. Curr. Biol. 5, 737–739 (1995)
Paterson, A. H. et al. The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 (2009)
The International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768 (2010)
Smith, D. B. & Flavell, R. B. Characterisation of the wheat genome by association genetics. Chromosoma 50, 223–242 (1975)
Baucom, R. S. et al. Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 5, e1000732 (2009)
Wicker, T. et al. Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives. Plant Cell 23, 1706–1718 (2011)
Choulet, F. et al. Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell 22, 1686–1701 (2010)
Gill, B. S. et al. A workshop report on wheat genome sequencing: International Genome Research on Wheat Consortium. Genetics 168, 1087–1096 (2004)
Paux, E. et al. A physical map of the 1-gigabase bread wheat chromosome 3B. Science 322, 101–104 (2008)
International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature 436, 793–800 (2005)
Li, L., Stoeckert, C. J., Jr & Roos, D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003)
Mochida, K., Yoshida, T., Sakurai, T., Ogihara, Y. & Shinozaki, K. TriFLDB: a database of clustered full-length coding sequences from Triticeae with applications to comparative grass genomics. Plant Physiol. 150, 1135–1146 (2009)
Richter, D. C., Ott, F., Auch, A. F., Schmid, R. & Huson, D. H. MetaSim: a sequencing simulator for genomics and metagenomics. PLoS ONE 3, e3373 (2008)
Hernandez, P. et al. Next-generation sequencing and syntenic integration of flow-sorted arms of wheat chromosome 4A exposes the chromosome structure and gene content. Plant J. 69, 377–386 (2012)
Massa, A. N. et al. Gene space dynamics during the evolution of Aegilops tauschii, Brachypodium distachyon, Oryza sativa, and Sorghum bicolor genomes. Mol. Biol. Evol. 28, 2537–2547 (2011)
The International Barley Genome Sequencing Consortium. A physical, genetic, and functional sequence assembly of the barley genome. Nature doi:10.1038/nature11543 (this issue).
Schnable, P. S. et al. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115 (2009)
Lee, E. K. et al. A functional phylogenomic view of the seed plants. PLoS Genet. 7, e1002411 (2011)
Allen, A. M. et al. Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol. J. 9, 1086–1099 (2011)
Salse, J. et al. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20, 11–24 (2008)
Qi, L. L. et al. A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168, 701–712 (2004)
Ozkan, H., Levy, A. A. & Feldman, M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13, 1735–1747 (2001)
Shavrukov, Y., Langridge, P. & Tester, M. Salinity tolerance and sodium exclusion in genus Triticum. Breed. Sci. 59, 671–678 (2009)
Wang, S., Yin, L., Tanaka, K., Tanaka, H. & Tsujimoto, H. Wheat-Aegilops chromosome addition lines showing high iron and zinc contents in grains. Breed. Sci. 61, 189–195 (2011)
Jiang, N., Bao, Z., Zhang, X., Eddy, S. R. & Wessler, S. R. Pack-MULE transposable elements mediate gene evolution in plants. Nature 431, 569–573 (2004)
Morgante, M. et al. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nature Genet. 37, 997–1002 (2005)
Jin, Y. K. & Bennetzen, J. L. Integration and nonrandom mutation of a plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell 6, 1177–1186 (1994)
Lippman, Z. et al. Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476 (2004)
Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000)
Schnable, J. C., Springer, N. M. & Freeling, M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl Acad. Sci. USA 108, 4069–4074 (2011)
Maere, S. et al. Modeling gene and genome duplications in eukaryotes. Proc. Natl Acad. Sci. USA 102, 5454–5459 (2005)
Gu, Y. Q., Coleman-Derr, D., Kong, X. & Anderson, O. D. Rapid genome evolution revealed by comparative sequence analysis of orthologous regions from four Triticeae genomes. Plant Physiol. 135, 459–470 (2004)
Mun, J. H. et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 10, R111 (2009)
Riley, R. Genetic control of cytologically diploid behaviour of hexaploid wheat. Nature 182, 713–715 (1958)
Buggs, R. J. et al. Rapid, repeated, and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Curr. Biol. 22, 248–252 (2012)
Pumphrey, M., Bai, J., Laudencia-Chingcuanco, D., Anderson, O. & Gill, B. S. Nonadditive expression of homoeologous genes is established upon polyploidization in hexaploid wheat. Genetics 181, 1147–1157 (2009)
Tester, M. & Langridge, P. Breeding technologies to increase crop production in a changing world. Science 327, 818–822 (2010)
Sears, E. R. in Chromosome Manipulation and Plant Genetics (eds Riley, R. & Lewis, K. R. ) 22–45 (Oliver and Boyd, 1966)
Acknowledgements
DNA sequence was generated by The University of Liverpool Centre for Genomic Research (United Kingdom), 454 Life Sciences (United States), The Cold Spring Harbor Woodbury Genome Centre (United States) and The Genome Analysis Centre (United Kingdom). This work was supported by UK Biological and Biotechnological Sciences Research Council (BBSRC) grants BB/G012865, BB/G013985/1 and BB/G013004/1, to K.J.E., M.W.B. and N. Hall; a Wolfson Merit Award from the Royal Society, to N. Hall; BBSRC Strategic Programme grant B/J004588/1 (GRO), to M.W.B.; EC TriticeaeGenome grant number 212019, to K.F.X.M. and M.W.B.; The TRITEX Project of the Plant20130 Initiative of the German Ministry of Education and Research grant number 0315954C, to K.F.X.M.; EC Transplant Grant 283496, to K.F.X.M. and P.K., a BBSRC Career Development Fellowship BB/H022333/1, to A.H., US NSF grants IOS-1032105 and DBI-0923128, to W.R.M.; USDA-NIFA grant 2008-35300-04588, to B.G.; and US NSF grants DBI-0701916, to J.D., and DBI-0822100, to S. Kianian.
Author information
Authors and Affiliations
Contributions
R.B., M.S., M.P., G.L.A.B. and R.D. are joint first authors. K.J.E., M.W.B., N. Hall and A.H. designed the project; W.R.M., M.K., M.T., I.B., J.D., M.-C.L., O.A., S. Kianian, N. Huo, B.G. and S.S. provided data and advice; R.D., N.M. and S. Kay conducted experiments; K.F.X.M., N. Hall and M.W.B. planned and conducted analyses; and R.B., M.S., M.P., G.L.A.B., A.M.A., D.B., D.W., P.K. and A.H. carried out analyses. K.J.E., A.H., W.R.M. and R.B. contributed to the text and M.W.B., N. Hall and K.F.X.M. wrote the manuscript. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
W.R.M. has participated in Illumina-sponsored meetings and received travel reimbursement and honoraria for presenting at these events.
Supplementary information
Supplementary Information
This file contains Supplementary Text and Data 1-5, Supplementary Tables 1-9, 13, 15-19 (see separate files for Supplementary Tables 10, 11, 12, 14), Supplementary Figures 1-6, 8-15 (see separate file for Supplementary Figure 7) and additional references. (PDF 1770 kb)
Supplementary Figure 7
This figure shows the separate maps of genetic markers on the A, B and D genomes. (PDF 1476 kb)
Supplementary Table 10
This table contains the over- and under-represented GO terms of expanded and contracted wheat gene families. (XLS 36 kb)
Supplementary Table 11
This table contains the over- and under-represented Pfam terms of expanded and contracted wheat gene families. (XLS 76 kb)
Supplementary Table 12
This table contains the over- and under-represented GO and Pfam terms of expanded Ae. tauschii gene families. (XLS 70 kb)
Supplementary Table 14
This table shows the Pfam domains in gene fragments. (XLS 205 kb)
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence (http://creativecommons.org/licenses/by-nc-sa/3.0/).
About this article
Cite this article
Brenchley, R., Spannagl, M., Pfeifer, M. et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491, 705–710 (2012). https://doi.org/10.1038/nature11650
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11650
This article is cited by
-
Characterization of sucrose nonfermenting-1-related protein kinase 2 (SnRK2) gene family in Haynaldia villosa demonstrated SnRK2.9-V enhances drought and salt stress tolerance of common wheat
BMC Genomics (2024)
-
Activation of the native PHYTOENE SYNTHASE 1 promoter by modifying near-miss cis-acting elements induces carotenoid biosynthesis in embryogenic rice callus
Plant Cell Reports (2024)
-
A high-resolution genotype–phenotype map identifies the TaSPL17 controlling grain number and size in wheat
Genome Biology (2023)
-
Identification of drought-tolerant hub genes in Iranian KC-2226 genotype of Aegilops tauschii using transcriptomic analysis
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.