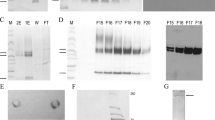
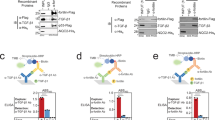
Abstract
Transforming growth factor (TGF)-β is stored in the extracellular matrix as a latent complex with its prodomain. Activation of TGF-β1 requires the binding of αv integrin to an RGD sequence in the prodomain and exertion of force on this domain, which is held in the extracellular matrix by latent TGF-β binding proteins. Crystals of dimeric porcine proTGF-β1 reveal a ring-shaped complex, a novel fold for the prodomain, and show how the prodomain shields the growth factor from recognition by receptors and alters its conformation. Complex formation between αvβ6 integrin and the prodomain is insufficient for TGF-β1 release. Force-dependent activation requires unfastening of a ‘straitjacket’ that encircles each growth-factor monomer at a position that can be locked by a disulphide bond. Sequences of all 33 TGF-β family members indicate a similar prodomain fold. The structure provides insights into the regulation of a family of growth and differentiation factors of fundamental importance in morphogenesis and homeostasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wu, M. Y. & Hill, C. S. TGF-β superfamily signaling in embryonic development and homeostasis. Dev. Cell 16, 329–343 (2009)
Derynck, R. & Miyazono, K. in The TGF-β Family (eds Derynck, R. & Miyazono, K. ) Ch. 2, 29–43 (Cold Spring Harbor Laboratory Press, 2008)
Blobe, G. C., Schiemann, W. P. & Lodish, H. F. Role of transforming growth factor β in human disease. N. Engl. J. Med. 342, 1350–1358 (2000)
Gray, A. M. & Mason, A. J. Requirement for activin A and transforming growth factor-β 1 pro-regions in homodimer assembly. Science 247, 1328–1330 (1990)
Annes, J. P., Chen, Y., Munger, J. S. & Rifkin, D. B. Integrin αVβ6-mediated activation of latent TGF-β requires the latent TGF-β binding protein-1. J. Cell Biol. 165, 723–734 (2004)
Ramirez, F. & Sakai, L. Y. Biogenesis and function of fibrillin assemblies. Cell Tissue Res. 339, 71–82 (2010)
Yang, Z. et al. Absence of integrin-mediated TGFβ1 activation in vivo recapitulates the phenotype of TGFβ1-null mice. J. Cell Biol. 176, 787–793 (2007)
Wipff, P. J. & Hinz, B. Integrins and the activation of latent transforming growth factor β1 — an intimate relationship. Eur. J. Cell Biol. 87, 601–615 (2008)
Aluwihare, P. et al. Mice that lack activity of αVβ6- and αVβ8-integrins reproduce the abnormalities of Tgfβ1- and Tgfβ3-null mice. J. Cell Sci. 122, 227–232 (2009)
Munger, J. S. et al. The integrin αVβ6 binds and activates latent TGF β1: A mechanism for regulating pulmonary inflammation and fibrosis. Cell 96, 319–328 (1999)
Yoshinaga, K. et al. Perturbation of transforming growth factor (TGF)-β1 association with latent TGF-β binding protein yields inflammation and tumors. Proc. Natl Acad. Sci. USA 105, 18758–18763 (2008)
Holm, L., Kaariainen, S., Rosenstrom, P. & Schenkel, A. Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 (2008)
Daopin, S., Piez, K. A., Ogawa, Y. & Davies, D. R. Crystal structure of transforming growth factor-β2: an unusual fold for the superfamily. Science 257, 369–373 (1992)
Schlunegger, M. P. & Grutter, M. G. An unusual feature revealed by the crystal structure at 2.2 Å resolution of human transforming growth factor-β2. Nature 358, 430–434 (1992)
Radaev, S. et al. Ternary complex of transforming growth factor-β1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily. J. Biol. Chem. 285, 14806–14814 (2010)
Gentry, L. E. & Nash, B. W. The pro domain of pre-pro-transforming growth factor β1 when independently expressed is a functional binding protein for the mature growth factor. Biochemistry 29, 6851–6857 (1990)
Belville, C. et al. Mutations of the anti-Müllerian hormone gene in patients with persistent Müllerian duct syndrome: biosynthesis, secretion, and processing of the abnormal proteins and analysis using a three-dimensional model. Mol. Endocrinol. 18, 708–721 (2004)
Walton, K. L. et al. Two distinct regions of latency-associated peptide coordinate stability of the latent transforming growth factor-β1 complex. J. Biol. Chem. 285, 17029–17037 (2010)
Little, S. C. & Mullins, M. C. Bone morphogenetic protein heterodimers assemble heteromeric type I receptor complexes to pattern the dorsoventral axis. Nature Cell Biol. 11, 637–643 (2009)
Lack, J. et al. Solution structure of the third TB domain from LTBP1 provides insight into assembly of the large latent complex that sequesters latent TGF-β. J. Mol. Biol. 334, 281–291 (2003)
Chen, Y. et al. Amino acid requirements for formation of the TGF-β-latent TGF-β binding protein complexes. J. Mol. Biol. 345, 175–186 (2005)
Luo, B.-H., Carman, C. V. & Springer, T. A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25, 619–647 (2007)
Forman, J. R. & Clarke, J. Mechanical unfolding of proteins: insights into biology, structure and folding. Curr. Opin. Struct. Biol. 17, 58–66 (2007)
Janssens, K. et al. Camurati-Engelmann disease: review of the clinical, radiological, and molecular data of 24 families and implications for diagnosis and treatment. J. Med. Genet. 43, 1–11 (2005)
Walton, K. L. et al. A common biosynthetic pathway governs the dimerization and secretion of inhibin and related transforming growth factor β (TGFβ) ligands. J. Biol. Chem. 284, 9311–9320 (2009)
Anderson, S. B., Goldberg, A. L. & Whitman, M. Identification of a novel pool of extracellular pro-myostatin in skeletal muscle. J. Biol. Chem. 283, 7027–7035 (2008)
Sengle, G. et al. Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J. Biol. Chem. 283, 13874–13888 (2008)
Cui, Y. et al. The activity and signaling range of mature BMP-4 is regulated by sequential cleavage at two sites within the prodomain of the precursor. Genes Dev. 15, 2797–2802 (2001)
Blanchet, M. H. et al. Cripto recruits Furin and PACE4 and controls Nodal trafficking during proteolytic maturation. EMBO J. 27, 2580–2591 (2008)
Wilson, C. A. et al. Müllerian inhibiting substance requires its N-terminal domain for maintenance of biological activity, a novel finding within the transforming growth factor-β superfamily. Mol. Endocrinol. 7, 247–257 (1993)
Ulloa, L. et al. Lefty proteins exhibit unique processing and activate the MAPK pathway. J. Biol. Chem. 276, 21387–21396 (2001)
Keefe, A. D., Wilson, D. S., Seelig, B. & Szostak, J. W. One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein Expr. Purif. 23, 440–446 (2001)
Zou, Z. & Sun, P. D. Overexpression of human transforming growth factor-β1 using a recombinant CHO cell expression system. Protein Expr. Purif. 37, 265–272 (2004)
Gentry, L. E. et al. Type 1 transforming growth factor beta: amplified expression and secretion of mature and precursor polypeptides in Chinese hamster ovary cells. Mol. Cell. Biol. 7, 3418–3427 (1987)
Brunner, A. M. et al. Site-directed mutagenesis of glycosylation sites in the transforming growth factor-beta 1 (TGF beta 1) and TGF beta 2 (414) precursors and of cysteine residues within mature TGF beta 1: effects on secretion and bioactivity. Mol. Endocrinol. 6, 1691–1700 (1992)
Heras, B. & Martin, J. L. Post-crystallization treatments for improving diffraction quality of protein crystals. Acta Crystallogr. D 61, 1173–1180 (2005)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Kabsch, W. in International Tables for Crystallography, Vol. F: Crystallography of Biological Macromolecules (eds Rossmann, M. G. & Arnold, E. V. ) Ch. 25.2.9 XDS, 730–734 (Springer, 2001)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D 66, 22–25 (2010)
Bailey, S. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Cowtan, K. Recent developments in classical density modification. Acta Crystallogr. D 66, 470–478 (2010)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
Takagi, J., Petre, B. M., Walz, T. & Springer, T. A. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 110, 599–611 (2002)
Chen, X. et al. Requirement of open headpiece conformation for activation of leukocyte integrin αXβ2 . Proc. Natl Acad. Sci. USA 107, 14727–14732 (2010)
Acknowledgements
We thank P. Sun for porcine TGF-β1 cDNA, K. Koli for human TGF-β1 and LTBP1 cDNAs, D. Rifkin for human LTBP1 cDNA and transformed mink lung epithelial cells, and the staff of the Advanced Photon Source General Medical Sciences and National Cancer Institute (APS GM/CA-CAT) beamline 23-ID.
Author information
Authors and Affiliations
Contributions
M.S. cloned, expressed and purified proTGF-β1, crystallized the protein, collected and processed X-ray data, refined and analysed the structure, designed and performed biochemical assays and wrote the paper. J.Z. collected and processed X-ray data, refined and analysed the structure and performed electron microscopy studies. R.W. designed and performed TGF-β1 assays. X.C. performed electron microscopy studies. L.M. processed X-ray data. T.W. provided electron microscopy supervision. T.A.S. designed and supervised the project, refined and analysed the structure and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-5 with legends and Supplementary Table 1. (PDF 7314 kb)
Rights and permissions
About this article
Cite this article
Shi, M., Zhu, J., Wang, R. et al. Latent TGF-β structure and activation. Nature 474, 343–349 (2011). https://doi.org/10.1038/nature10152
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10152
This article is cited by
-
The roles and regulatory mechanisms of TGF-β and BMP signaling in bone and cartilage development, homeostasis and disease
Cell Research (2024)
-
Fibroblast and myofibroblast activation in normal tissue repair and fibrosis
Nature Reviews Molecular Cell Biology (2024)
-
Regulatory T cell-mediated immunosuppression orchestrated by cancer: towards an immuno-genomic paradigm for precision medicine
Nature Reviews Clinical Oncology (2024)
-
Emerging roles for the ADAMTS-like family of matricellular proteins in cardiovascular disease through regulation of the extracellular microenvironment
Molecular Biology Reports (2024)
-
TGF-β1 signalling in Alzheimer’s pathology and cytoskeletal reorganization: a specialized Tau perspective
Journal of Neuroinflammation (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.