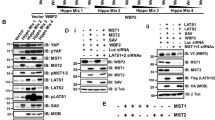
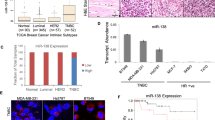
Abstract
Aberrant expression of microRNAs (miRNAs) and the enzymes that control their processing have been reported in multiple biological processes including primary and metastatic tumours1,2,3,4,5,6, but the mechanisms governing this are not clearly understood. Here we show that TAp63, a p53 family member, suppresses tumorigenesis and metastasis, and coordinately regulates Dicer and miR-130b to suppress metastasis. Metastatic mouse and human tumours deficient in TAp63 express Dicer at very low levels, and we found that modulation of expression of Dicer and miR-130b markedly affected the metastatic potential of cells lacking TAp63. TAp63 binds to and transactivates the Dicer promoter, demonstrating direct transcriptional regulation of Dicer by TAp63. These data provide a novel understanding of the roles of TAp63 in tumour and metastasis suppression through the coordinate transcriptional regulation of Dicer and miR-130b and may have implications for the many processes regulated by miRNAs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
26 May 2023
Editor’s Note: Readers are alerted that the reliability of data presented in this manuscript is currently in question. Appropriate editorial action will be taken once this matter is resolved.
References
Baffa, R. et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J. Pathol.219, 214–221 (2009)
Gibbons, D. L. et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev.23, 2140–2151 (2009)
Ma, L., Teruya-Feldstein, J. & Weinberg, R. A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature449, 682–628 (2007)
Kumar, M. S. et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev.23, 2700–2704 (2009)
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature435, 834–8 (2005)
Merritt, W. M. et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med.359, 2641–2650 (2008)
Flores, E. R. The roles of p63 in cancer. Cell Cycle6, 300–304 (2007)
Flores, E. R. et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell7, 363–373 (2005)
Keyes, W. M. et al. p63 heterozygous mutant mice are not prone to spontaneous or chemically induced tumors. Proc. Natl Acad. Sci. USA103, 8435–8440 (2006)
Yang, A., Kaghad, M., Caput, D. & McKeon, F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet.18, 90–95 (2002)
Hibi, K. et al. AIS is an oncogene amplified in squamous cell carcinoma. Proc. Natl Acad. Sci. USA97, 5462–5467 (2000)
Park, B. J. et al. Frequent alteration of p63 expression in human primary bladder carcinomas. Cancer Res.60, 3370–3374 (2000)
Urist, M. J. et al. Loss of p63 expression is associated with tumor progression in bladder cancer. Am. J. Pathol.161, 1199–1206 (2002)
Park, H. R. et al. Low expression of p63 and p73 in osteosarcoma. Tumori90, 239–243 (2004)
Yang, A. et al. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell2, 305–316 (1998)
Jacks, T. et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol.4, 1–7 (1994)
Lang, G. A. et al. Gain of function of a p53 hot spot mutation in a mouse model of Li–Fraumeni syndrome. Cell119, 861–872 (2004)
Olive, K. P. et al. Mutant p53 gain of function in two mouse models of Li–Fraumeni syndrome. Cell119, 847–860 (2004)
Su, X. et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell5, 64–75 (2009)
Maruya, S. et al. Differential methylation status of tumor-associated genes in head and neck squamous carcinoma: incidence and potential implications. Clin. Cancer Res.10, 3825–3830 (2004)
Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev.15, 3243–3248 (2001)
Kumar, M. S. et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nature Genet.39, 673–677 (2007)
He, L. et al. A microRNA component of the p53 tumour suppressor network. Nature447, 1130–1134 (2007)
Flores, E. R. et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature416, 560–564 (2002)
Pruneri, G. et al. The transactivating isoforms of p63 are overexpressed in high-grade follicular lymphomas independent of the occurrence of p63 gene amplification. J. Pathol.206, 337–345 (2005)
Mudhasani, R. et al. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J. Cell Biol.181, 1055–1063 (2008)
Su, X. et al. Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf . EMBO J.28, 1904–1915 (2009)
Landthaler, M. et al. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA14, 2580–2596 (2008)
Lin, Y. L. et al. p63 and p73 transcriptionally regulate genes involved in DNA repair. PLoS Genet.5, e1000680 (2009)
Acknowledgements
We thank C.-g. Liu for technical assistance with miRNA analyses and A. Multani for metaphase analyses (funded by NCI CA16672); K. Y. Tsai for scientific discussions and comments on the manuscript; and D. Yu, J. Jackson and S. Rehman for reagents and advice. This work was supported by grants to E.R.F. from the American Cancer Society (RSG-07-082-01-MGO), the Susan G. Komen Foundation (BCTR600208) and the Hildegardo E. and Olga M. Flores Foundation. This work was supported in part by an NCI-Cancer Center Core Grant (CA-16672) (University of Texas M. D. Anderson Cancer Center), a Career Development Award from the Genitourinary Cancer SPORE (P50CA091846) to E.R.F., Lung Cancer SPORE (P50CA070907) to I.W. and U01DE019765 to A.E.-N. E.R.F. is a scholar of the Rita Allen Foundation and the V Foundation for Cancer Research.
Author information
Authors and Affiliations
Contributions
X.S., D.C. and E.R.F. designed the experiments and analysed the data. X.S., D.C., M.S.C., L.L., Y.J.G., Y.-L.L. and M.L.L. performed the experiments. A.E.-N. performed pathology analysis and provided human HNSCC samples. M.B.S. and I.W. provided human lung adenocarcinoma pathology information and RNA samples. C.J.C. performed statistical analysis on miRNA data. X.S., D.C. and E.R.F. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-7 with legends and Supplementary Tables 1- 4. (PDF 12105 kb)
Rights and permissions
About this article
Cite this article
Su, X., Chakravarti, D., Cho, M. et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 467, 986–990 (2010). https://doi.org/10.1038/nature09459
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09459
This article is cited by
-
microRNA-205 represses breast cancer metastasis by perturbing the rab coupling protein [RCP]-mediated integrin β1 recycling on the membrane
Apoptosis (2024)
-
Identification of multiple organ metastasis-associated hub mRNA/miRNA signatures in non-small cell lung cancer
Cell Death & Disease (2023)
-
STAT3/miR-130b-3p/MBNL1 feedback loop regulated by mTORC1 signaling promotes angiogenesis and tumor growth
Journal of Experimental & Clinical Cancer Research (2022)
-
p63, a key regulator of Ago2, links to the microRNA-144 cluster
Cell Death & Disease (2022)
-
Structural diversity of p63 and p73 isoforms
Cell Death & Differentiation (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.