Abstract
Soybean (Glycine max) is one of the most important crop plants for seed protein and oil content, and for its capacity to fix atmospheric nitrogen through symbioses with soil-borne microorganisms. We sequenced the 1.1-gigabase genome by a whole-genome shotgun approach and integrated it with physical and high-density genetic maps to create a chromosome-scale draft sequence assembly. We predict 46,430 protein-coding genes, 70% more than Arabidopsis and similar to the poplar genome which, like soybean, is an ancient polyploid (palaeopolyploid). About 78% of the predicted genes occur in chromosome ends, which comprise less than one-half of the genome but account for nearly all of the genetic recombination. Genome duplications occurred at approximately 59 and 13 million years ago, resulting in a highly duplicated genome with nearly 75% of the genes present in multiple copies. The two duplication events were followed by gene diversification and loss, and numerous chromosome rearrangements. An accurate soybean genome sequence will facilitate the identification of the genetic basis of many soybean traits, and accelerate the creation of improved soybean varieties.
Similar content being viewed by others
Main
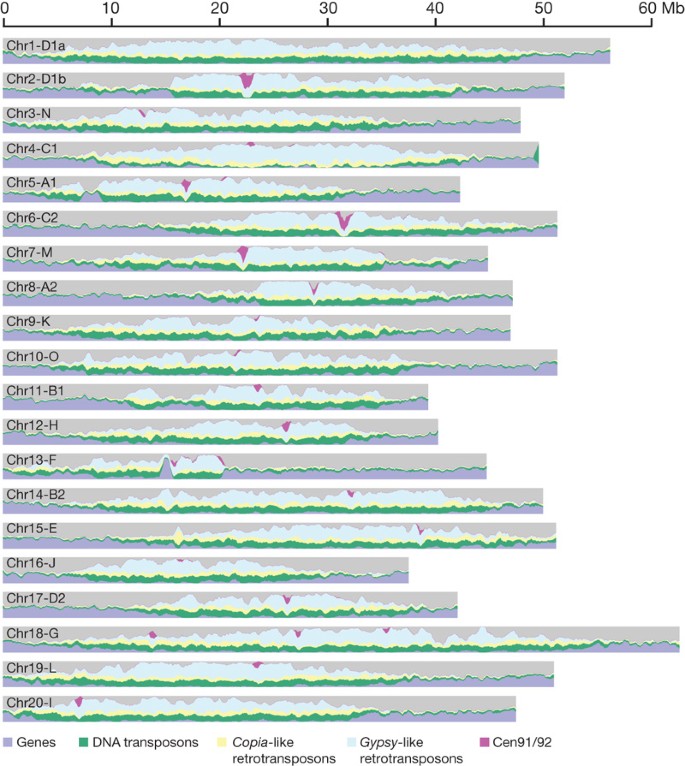
Legumes are an important part of world agriculture as they fix atmospheric nitrogen by intimate symbioses with microorganisms. The soybean in particular is important worldwide as a predominant plant source of both animal feed protein and cooking oil. We report here a soybean whole-genome shotgun sequence of Glycine max var. Williams 82, comprised of 950 megabases (Mb) of assembled and anchored sequence (Fig. 1), representing about 85% of the predicted 1,115-Mb genome1 (Supplementary Table 3.1). Most of the genome sequence (Fig. 1) is assembled into 20 chromosome-level pseudomolecules containing 397 sequence scaffolds with ordered positions within the 20 soybean linkage groups. An additional 17.7 Mb is present in 1,148 unanchored sequence scaffolds that are mostly repetitive and contain fewer than 450 predicted genes. Scaffold placements were determined with extensive genetic maps, including 4,991 single nucleotide polymorphisms (SNPs) and 874 simple sequence repeats (SSRs)2,3,4,5. All but 20 of the 397 sequence scaffolds are unambiguously oriented on the chromosomes. Unoriented scaffolds are in repetitive regions where there is a paucity of recombination and genetic markers (see Supplementary Information for assembly details).
Major DNA components are categorized into genes (blue), DNA transposons (green), Copia-like retrotransposons (yellow), Gypsy-like retrotransposons (cyan) and Cent91/92 (a soybean-specific centromeric repeat (pink)), with respective DNA contents of 18%, 17%, 13%, 30% and 1% of the genome sequence. Unclassified DNA content is coloured grey. Categories were determined for 0.5-Mb windows with a 0.1-Mb shift.
The soybean genome is the largest whole-genome shotgun-sequenced plant genome so far and compares favourably to all other high-quality draft whole-genome shotgun-sequenced plant genomes (Supplementary Table 4). A total of 8 of the 20 chromosomes have telomeric repeats (TTTAGGG or CCCTAAA) on both of the distal scaffolds and 11 other chromosomes have telomeric repeats on a single arm, for a total of 27 out of 40 chromosome ends captured in sequence scaffolds. Also, internal scaffolds in 19 of 20 chromosomes contain a large block of characteristic 91- or 92-base-pair (bp) centromeric repeats6,7 (Fig. 1). Four chromosome assemblies contain several 91/92-bp blocks; this may be the correct physical placements of these sequences, or may reflect the difficulty in assembling these highly repetitive regions.
Gene composition and repetitive DNA
A striking feature of the soybean genome is that 57% of the genomic sequence occurs in repeat-rich, low-recombination heterochromatic regions surrounding the centromeres. The average ratio of genetic-to-physical distance is 1 cM per 197 kb in euchromatic regions, and 1 cM per 3.5 Mb in heterochromatic regions (see Supplementary Information section 1.8). For reference, these proportions are similar to those in Sorghum, in which 62% of the sequence is heterochromatic, and different than in rice, with 15% in heterochromatin8. In general, these boundaries, determined on the basis of suppressed recombination, correlate with transitions in gene density and transposon density. Ninety-three per cent of the recombination occurs in the repeat-poor, gene-rich euchromatic genomic region that only accounts for 43% of the genome. Nevertheless, 21.6% of the high-confidence genes are found in the repeat- and transposon-rich regions in the chromosome centres.
We identified 46,430 high-confidence protein-coding loci in the soybean genome, using a combination of full-length complementary DNAs9, expressed sequence tags, homology and ab initio methods (Supplementary Information section 2). Another ∼20,000 loci were predicted with lower confidence; this set is enriched for hypothetical, partial and/or transposon-related sequences, and possess shorter coding sequences and fewer introns than the high-confidence set. The exon–intron structure of genes shows high conservation among soybean, poplar and grapevine, consistent with a high degree of position and phase conservation found more broadly across angiosperms10. Introns in soybean gene pairs retained in duplicate have a strong tendency to persist. Of 19,775 introns shared by poplar and grapevine (diverged more than 90 million years (Myr) ago11), and hence by the last common ancestor of soybean and grapevine, 19,666 (99.45%) were preserved in both copies in soybean. Of the remaining 0.55%, 78% are absent in both recent soybean copies (that is, lost before the ∼13-Myr-ago duplication) and 22% are found only in one paralogue (that is, other copy lost). We find a slower intron loss rate in poplar (0.4%) than in soybean (0.6%) since the last common rosid ancestor, which is consistent with the slower rate of sequence evolution in the poplar lineage thought to be associated with its perennial, clonal habit, global distribution and wind pollination12. Intron size is also highly conserved in recent soybean paralogues, indicating that few insertions and deletions have accumulated within introns over the past 13 Myr.
Of the 46,430 high-confidence loci, 34,073 (73%) are clearly orthologous with one or more sequences in other angiosperms, and can be assigned to 12,253 gene families (Supplementary Table 5). Among pan-angiosperm or pan-rosid gene families that also have members outside the legumes, soybean is particularly enriched (using a Fisher’s exact test relative to Arabidopsis) in genes containing NB-ARC (nucleotide-binding-site-APAF1-R-Ced) and LRR (leucine-rich-repeat) domains. These genes are associated with the plant immune system, and are known to be dynamic13. Tandem gene family expansions are common in soybean and include NBS-LRR, F-box, auxin-responsive protein, and other domains commonly found in large gene families in plants. The ages of genes in these tandem families, inferred from intrafamily sequence divergence, indicate that they originated at various times in the evolutionary history of soybean, rather than in a discrete burst.
From protein families in the sequenced angiosperms (http://www.phytozome.net) (Supplementary Table 4), we identified 283 putative legume-specific gene families containing 448 high-confidence soybean genes (Supplementary Information section 2). These gene families include soybean and Medicago representatives, but no representatives from grapevine, poplar, Arabidopsis, papaya, or grass (Sorghum, rice, maize, Brachypodium). The top domains in this set are the AP2 domain, protein kinase domain, cytochrome P450, and PPR repeat. An additional 741 putatively soybean-specific gene families (each consisting of two or more high-confidence soybean genes) may also include legume-specific genes that have not yet been sequenced in the ongoing Medicago sequencing project, or may represent bona fide soybean-specific genes. The top domains in this list include protein kinase and protein tyrosine kinase, AP2, LRR, MYB-like DNA binding domain, cytochrome P450 (the same domains most common in the entire soybean proteome) as well as GDSL-like lipase/acylhydrolase and stress-upregulated Nod19.
A combination of structure-based analyses and homology-based comparisons resulted in identification of 38,581 repetitive elements, covering most types of plant transposable elements. These elements, together with numerous truncated elements and other fragments, make up ∼59% of the soybean genome (Supplementary Table 6).
Long terminal repeat (LTR) retrotransposons are the most abundant class of transposable elements. The soybean genome contains ∼42% LTR retrotransposons, fewer than Sorghum8 and maize14, but higher than rice15. The intact element sizes range from 1 kb to 21 kb, with an average size of 8.7 kb (Supplementary Fig. 2). Of the 510 families containing 14,106 intact elements, 69% are Gypsy-like and the remainder Copia-like. However, most (∼78%) of these families are present at low copy numbers, typically fewer than 10 copies. The genome also contains an estimated 18,264 solo LTRs, probably caused by homologous recombination between LTRs from a single element. Nested retrotransposons are common, with 4,552 nested insertion events identified. The copy numbers within each block range from one to six.
The genome consists of ∼17% transposable elements, divided into Tc1/Mariner, haT, Mutator, PIF/Harbinger, Pong, CACTA superfamilies and Helitrons. Of these superfamilies, those containing more than 65 complete copies, Tc1/Mariner and Pong, comprise ∼0.1% of the genome sequence, and seem to have not undergone recent amplification, indicating that they may be inactive and relatively old. Conversely, other families seem to have amplified recently and may still be active, indicated by the high similarity (>98%) of multiple elements.
Multiple whole-genome duplication events
Timing and phylogenetic position
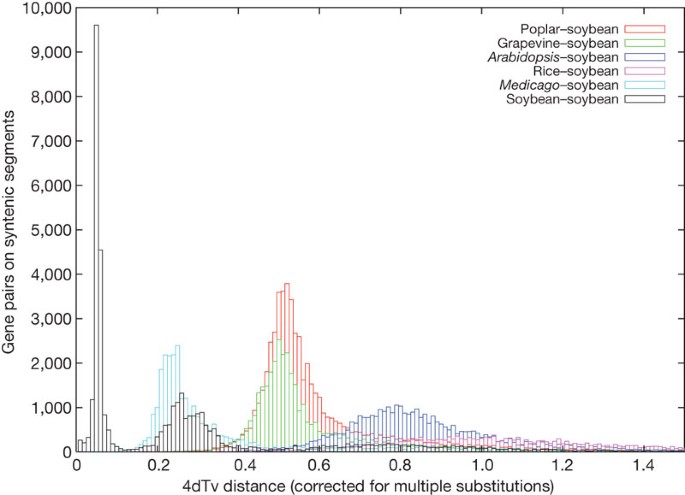
A striking feature of the soybean genome is the extent to which blocks of duplicated genes have been retained. On the basis of previous studies that examined pairwise synonymous distance (Ks values) of paralogues16,17, and targeted sequencing of duplicated regions within the soybean genome18, we expected that large homologous regions would be identified in the genome. Using a pattern-matching search, gene families of sizes from two to six were identified, and Ks values were calculated for these genes, here displayed as a histogram plot (Fig. 2), which shows two distinct peaks. Similarly, nucleotide diversity for the fourfold synonymous third-codon transversion position, 4dTv, was calculated. Both metrics give a measure of divergence between two genes, but the 4dTv uses a subset of the sites (transitions/transversion) used in the computation of Ks. 31,264 high-confidence soybean genes have recent paralogues with Ks ≈ 0.13 synonymous substitutions per site and 4dTv ≈ 0.0566 synonymous transversions per site (Fig. 3), corresponding to a soybean-lineage-specific palaeotetraploidization. This was probably an allotetraploidy event based on chromosomal evidence19. Of the 46,430 high-confidence genes, 31,264 exist as paralogues and 15,166 have reverted to singletons. We infer that the pre-duplication proto-soybean genome possessed ∼30,000 genes: half of (2 × 15,166 + 2 × 15,632) = 30,798. This number is comparable to the modern Arabidopsis gene complement. A second paralogue peak at Ks ≈ 0.59 (4dTv ≈ 0.26) corresponds to the early-legume duplication, which several lines of evidence suggest occurred near the origins of the papilionoid lineage20. The papilionoid origin has been dated to approximately 59 Myr ago21. A third highly diffuse peak is seen when the plot is expanded past a Ks value of 1.5 (data not shown) and most probably corresponds to the ‘gamma’ event22, shown to be a triplication in Vitis23 and in other angiosperms24.
The bottom histogram plot shows pairwise Ks values for gene family sizes 2 to 6. Top panels show the 20 chromosomes in a circle with lines connecting homologous genes. Gene-rich regions (euchromatin) of each chromosome are coded a different colour around the circle. Grey represents Ks values of 0.06–0.39, 13-Myr genome duplication; black represents Ks values of 0.40–0.80, 59-Myr genome duplication. These correspond to the grey and black bars in the histogram. a, Chromosomes 1, 11, 17, 7, 10 and 3, which contain centromeric repeat Sb91. b, Chromosomes 19 and 6, which contain both Sb91 and Sb92 centromeric repeats. c, Chromosomes 18, 5, 8, 2, 14, 12, 13, 9, 15, 16, 20 and 4, which contain Sb92.
Segments were found by locating blocks of BLAST hits with significance 1×10-18 or better with less than 10 intervening genes between such hits. The 4dTv distance between orthologous genes on these segments is reported.
Owing to the existence of macrofossils in the legumes and allies, the timing of clade origins in the legumes is better established than other plant families. A fossil-calibrated molecular clock for the legumes places the origin of the legume stem clade and the oldest papilionoid crown clade at 58 to 60 Myr ago21. If the early-legume whole-genome duplication (WGD) occurred outside the papilionoid lineage, as suggested by map evidence from Arachis (an early-diverging genus in the papilionoid clade)20, then the duplication occurred within the narrow window of time between the origin of the legumes and the papilionoid radiation. If the older duplication is assumed to have occurred around 58 Myr ago, then the calculated rate of silent mutations extending back to the duplication would be 5.17 × 10-3, similar to previous estimates of 5.2 × 10–3 (ref. 21). The Glycine-specific duplication is estimated to have occurred ∼13 Myr ago, an age consistent with previous estimates16,17.
Structural organization
We identified homologous blocks within the genome using i-ADHoRe25. Using relatively stringent parameters, 442 multiplicons (that is, duplicated segments) were identified within the soybean genome and visualized using Circos26 (Fig. 2). Owing to the multiple rounds of duplication and diploidization in the genome, as well as chromosomal rearrangements, multiplicons (or blocks) between chromosomes can involve more than just two chromosomes. On average, 61.4% of the homologous genes are found in blocks involving only two chromosomes, only 5.63% spanning three chromosomes, and 21.53% traversing four chromosomes. Two notable exceptions to this pattern are chromosome 14, which has 11.8% of its genes retained across three chromosomes, and chromosome 20 with 7.08% of the homologues (gene pairs resulting from genome duplication) retained across four chromosomes. Chromosome 14 seems to be a highly fragmented chromosome with block matches to 14 other chromosomes, the highest number of all chromosomes. Conversely, chromosome 20 is highly homologous to the long arm of chromosome 10, with few matches elsewhere in the genome.
Retention of homologues across the genome is exceptionally high; blocks retained in two or more chromosomes can be clearly observed (Fig. 2 and Supplementary Figs 5 and 6). The number of homologues (gene pairs) within a block average 31, although any given block may contain from 6 to 736 homologues. Given that not all genes within a block are retained as homologues (owing to loss of duplicated genes over time (fractionation)), the average number of genes in a block is ∼75 genes and ranges from 8 to 1,377 genes.
Repeated duplications in the soybean genome make it possible to determine rates of gene loss following each round of polyploidy. In homologous segments from the 13-Myr-old Glycine duplication, 43.4% of genes have matches in the corresponding region, in contrast to 25.9% in blocks from the early legume duplication. Combining these gene-loss rates with WGD dates of 13 Myr ago and 59 Myr ago, the rate of gene loss has been 4.36% of genes per Myr following the Glycine WGD and 1.28% of genes per Myr following the early-legume WGD. This differential in gene-loss rates indicates an exponential decay pattern of rapid gene loss after duplication, slowing over time.
Nodulation and oil biosynthesis genes
A unique feature of legumes is their ability to establish nitrogen-fixing symbioses with soil bacteria of the family Rhizobiaceae. Therefore, information on the nodulation functions of the soybean genome is of particular interest. Sequence comparisons with previously identified nodulation genes identified 28 nodulin genes and 24 key regulatory genes, which probably represent true orthologues of known nodulation genes in other legume species (Supplementary section 3 and Supplementary Table 8). Among this list of 52 genes, 32 have at least one highly conserved homologue gene. We hypothesize that these are homologous gene pairs arising from the Glycine WGD (that is, ∼13 Myr ago). Further analysis shows that seven soybean nodulin genes produce transcript variants. The exceptional example is nodulin-24 (Glyma14g05690), which seems to produce ten transcript variants (Supplementary Table 8). In total, 25% of the examined nodulin genes produce transcript variants, which is slightly higher than the incidence of alternative splicing in Arabidopsis (∼21.8%) and rice (∼21.2%)27. However, none of the soybean regulatory nodulation genes produces transcript variants (Supplementary Table 8).
Mining the soybean genome for genes governing metabolic steps in triacylglycerol biosynthesis could prove beneficial in efforts to modify soybean oil composition or content. Genomic analysis of acyl lipid biosynthesis in Arabidopsis revealed 614 genes involved in pathways beginning with plastid acetyl-CoA production for de novo fatty acid synthesis through cuticular wax deposition28. Comparison of these sequences to the soybean genome identified 1,127 putative orthologous and paralogous genes in soybean. This is probably a low estimate owing to the high stringency conditions used for gene mining. The distribution of these genes according to various functional classes of acyl lipid biosynthesis is shown in Table 1. Comparing Arabidopsis to soybean, the number of genes involved in storage lipid synthesis, fatty acid elongation and wax/cutin production was similar. For all other subclasses, the soybean genome contained substantially higher numbers of genes. Interestingly, the number of genes involved in lipid signalling, degradation of storage lipids, and membrane lipid synthesis were two- to threefold higher in soybean than Arabidopsis, indicating that these areas of acyl lipid synthesis are more complex in soybean. The number of genes involved in plastid de novo fatty acid synthesis was 63% higher in soybean compared to Arabidopsis. Many single-gene activities in Arabidopsis are encoded by multigene families in soybean, including ketoacyl-ACP synthase II (12 copies in soybean), malonyl-CoA:ACP malonyltransferase (2 copies), enoyl-ACP reductase (5 copies), acyl-ACP thioesterase FatB (6 copies) and plastid homomeric acetyl-CoA carboxylase (3 copies). Long-chain acyl-CoA synthetases, ER acyltransferases, mitochondrial glycerol-phosphate acyltransferases, and lipoxygenases are all unusually large gene families in soybean, containing as many as 24, 21, 20 and 52 members, respectively. The multigenic nature of these and many other activities involved in acyl lipid metabolism suggests the potential for more complex transcriptional control in soybean compared to Arabidopsis.
Transcription factor diversity
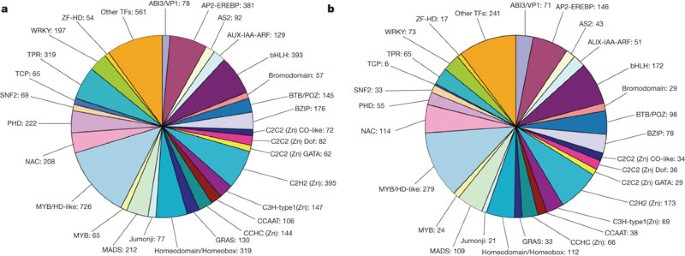
We identified soybean transcription factor genes by sequence comparison to known transcription factor gene families, as well as by searching for known DNA-binding domains. In total, 5,671 putative soybean transcription factor genes, distributed in 63 families, were identified (Fig. 4a and Supplementary Table 9). This number represents 12.2% of the 46,430 predicted soybean protein-coding loci. A similar analysis performed on the Arabidopsis genome identified 2,315 putative Arabidopsis transcription factor genes, representing 7.1% of the 32,825 predicted Arabidopsis protein-coding loci (Fig. 4b). Transcription factor genes are homogeneously distributed across the chromosomes in both soybean and Arabidopsis, with an average relative abundance of 8–10% transcription factor genes on each chromosome. On rare occasions, regions were identified in both genomes that had a relatively low (<5%) or high density (>12%) of transcription factor genes. Among the transcription factor genes identified, 9.5% of soybean genes (538 transcription factor genes) and 8.2% of Arabidopsis genes (190 Arabidopsis transcription factor genes) are tandemly duplicated. By way of example, only one region in Arabidopsis has more than five duplicated transcription factor genes in tandem (seven ABI3/VP1 genes (At4G31610 to At4G31660)), whereas in soybean several such regions are present (for example, 13 C3H-type 1 (Zn) (Glyma15g19120 to Glyma15g19240); six MYB/HD-like (Glyma06g45520 to Glyma06g45570); and five MADS (Glyma20g27320 to Glyma20g27360); Supplementary Table 8). The overall distribution of soybean transcription factor genes among the various known protein families is very similar between Arabidopsis and soybean (Supplementary Fig. 10a, b). However, some families are relatively sparser or more abundant in soybean, perhaps reflecting differences in biological function. For example, members of the ABI3/VP1 family are 2.2-times more abundant in Arabidopsis, whereas members of the TCP family are 4.4-times more abundant in soybean. In addition, those gene families with fewer members are differentially represented between soybean and Arabidopsis. FHA, HD-Zip (homeodomain/leucine zipper), PLATZ, SRS and TUB transcription factor genes are more abundant in soybean (2.7, 2.9, 4.1, 3, and 4.9 times, respectively) and HTH-ARAC (helix–turn–helix araC/xylS-type) genes were identified exclusively in soybean. In contrast, HSF, HTH-FIS (helix–turn–helix-factor for inversion stimulation), TAZ and U1-type (Zn) genes are present in relatively larger numbers in Arabidopsis (5.4, 4.9, 24.5 and 2.9 times, respectively). Notably, both ABI3/VP1, TCP, SRS and Tubby transcription factor genes were shown to have critical roles in plant development (for example, ABI3/VP1 during seed development; TCP, SRS and Tubby affect overall plant development29,30,31,32,33). The differences seen in relative transcription factor gene abundance indicates that regulatory pathways in soybean may differ from those described in Arabidopsis.
Only the distribution of the most representative transcription families is detailed here. AUX-IAA-ARF, indole-3-acetic acid-auxin response factor; BTB/POZ, bric-à-brac tramtrack broad complex/pox viruses and zinc fingers; BZIP, basic leucine zipper; GRAS, (GAI, RGA, SCR); NAC, (NAM, ATAF1/2, CUC2); PHD, plant homeodomain-finger transcription factor; TCP, (TB1, CYC, PCF); TFs, transcription factors; TPR, tetratricopepitide repeat; WRKY, conserved amino acid sequence WRKYGQK at its N-terminal end.
Impact on agriculture
Hundreds of qualitatively inherited (single gene) traits have been characterized in soybean and many genetically mapped. However, most important crop production traits and those important to seed quality for human health, animal nutrition and biofuel production are quantitatively inherited. The regions of the genome containing DNA sequence affecting these traits are called quantitative trait loci (QTL). QTL mapping studies have been ongoing for more than 90 distinct traits of soybean including plant developmental and reproductive characters, disease resistance, seed quality and nutritional traits. In most cases, the causal functional gene or transcription factor underlying the QTL is unknown. However, the integration of the whole genome sequence with the dense genetic marker map that now exists in soybean2,3,4,5 (http://www.Soybase.org) will allow the association of mapped phenotypic effectors with the causal DNA sequence. There are already examples where the availability of the soybean genomic sequence has accelerated these discovery efforts. Having access to the sequence allowed cloning and identification of the rsm1 (raffinose synthase) mutation that can be used to select for low-stachyose-containing soybean lines that will improve the ability of animals and humans to digest soybeans34. Using a comparative genomics approach between soybean and maize, a single-base mutation was found that causes a reduction in phytate production in soybean35. Phytate reduction could result in a reduction of a major environmental runoff contaminant from swine and poultry waste. Perhaps most exciting for the soybean community, the first resistance gene for the devastating disease Asian soybean rust (ASR) has been cloned with the aid of the soybean genomic sequence and confirmed with viral-induced gene silencing36. In countries where ASR is well established, soybean yield losses due to the disease can range from 10% to 80%36 and the development of soybean strains resistant to ASR will greatly benefit world soybean production.
Soybean, one of the most important global sources of protein and oil, is now the first legume species with a complete genome sequence. It is, therefore, a key reference for the more than 20,000 legume species, and for the remarkable evolutionary innovation of nitrogen-fixing symbiosis. This genome, with a common ancestor only 20 million years removed from many other domesticated bean species, will allow us to knit together knowledge about traits observed and mapped in all of the beans and relatives. The genome sequence is also an essential framework for vast new experimental information such as tissue-specific expression and whole-genome association data. With knowledge of this genome’s billion-plus nucleotides, we approach an understanding of the plant’s capacity to turn carbon dioxide, water, sunlight and elemental nitrogen and minerals into concentrated energy, protein and nutrients for human and animal use. The genome sequence opens the door to crop improvements that are needed for sustainable human and animal food production, energy production and environmental balance in agriculture worldwide.
Methods Summary
Seeds from cultivar Williams 82 were grown in a growth chamber for 2 weeks and etiolated for 5 days before harvest. A standard phenol/chloroform leaf extraction was performed. DNA was treated with RNase A and proteinase K and precipitated with ethanol.
All sequencing reads were collected with Sanger sequencing protocols on ABI 3730XL capillary sequencing machines, a majority at the Joint Genome Institute in Walnut Creek, California.
A total of 15,332,163 sequence reads were assembled using Arachne v.20071016 (ref. 37) to form 3,363 scaffolds covering 969.6 Mb of the soybean genome. The resulting assembly was integrated with the genetic and physical maps previously built for soybean and a newly constructed genetic map to produce 20 chromosome-scale scaffolds covering 937.3 Mb and an additional 1,148 unmapped scaffolds that cover 17.7 Mb of the genome.
Genes were annotated using Fgenesh+38 and GenomeScan39 informed by EST alignments and peptide matches to genome from Arabidopsis, rice and grapevine. Models were reconciled with EST alignments and UTR added using PASA40. Models were filtered for high confidence by penalizing genes which were transposable-element-related, had low sequence entropy, short introns, incomplete start or stop, low C-score, no UniGene hit at 1 × 10-5, or the model was less than 30% the length of its best hit.
LTR retrotransposons were identified by the program LTR_STRUC41, manually inspected to check structure features and classified into distinct families based on the similarities to LTR sequences. DNA transposons were identified using conserved protein domains as queries in TBLASTN42 searches of the genome. Identified elements were used as a custom library for RepeatMasker (current version: open 3.2.8; http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) to detect missed intact elements, truncated elements and fragments.
Virtual suffix trees with six-frame translation were generated using Vmatch43 and then clustered into families. Pairwise alignments between gene family members were performed using ClustalW44. Identification of homologous blocks was performed using i-ADHoRe v2.1 (ref. 25). Visualization of blocks was performed with Circos26.
Accession codes
Primary accessions
DDBJ/GenBank/EMBL
Data deposits
This whole-genome shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession ACUP00000000. The version described here is the first version, ACUP01000000. Full annotation is available at http://www.phytozome.net.
References
Arumuganathan, K. & Earle, E. D. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218 (1991)
Choi, I. Y. et al. A soybean transcript map: gene distribution, haplotype and single-nucleotide polymorphism analysis. Genetics 176, 685–696 (2007)
Hyten, D. L. et al. High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genomics (in the press)
Hyten, D. L. et al. A high density integrated genetic linkage map of soybean and the development of a 1,536 Universal Soy Linkage Panel for QTL mapping. Crop Sci. (in the press)
Song, Q. J. et al. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 109, 122–128 (2004)
Lin, J. Y. et al. Pericentromeric regions of soybean (Glycine max L. Merr.) chromosomes consist of retroelements and tandemly repeated DNA and are structurally and evolutionarily labile. Genetics 170, 1221–1230 (2005)
Vahedian, M. et al. Genomic organization and evolution of the soybean SB92 satellite sequence. Plant Mol. Biol. 29, 857–862 (1995)
Paterson, A. H. et al. The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 (2009)
Umezawa, T. et al. Sequencing and analysis of approximately 40,000 soybean cDNA clones from a full-length-enriched cDNA library. DNA Res. 15, 333–346 (2008)
Roy, S. W. & Penny, D. Patterns of intron loss and gain in plants: intron loss-dominated evolution and genome-wide comparison of O. sativa and A. thaliana . Mol. Biol. Evol. 24, 171–181 (2007)
Wang, H. et al. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc. Natl Acad. Sci. USA 106, 3853–3858 (2009)
Tuskan, G. A. et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 (2006)
Michelmore, R. & Meyers, B. C. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130 (1998)
Bruggmann, R. et al. Uneven chromosome contraction and expansion in the maize genome. Genome Res. 16, 1241–1251 (2006)
Ma, J., Devos, K. M. & Bennetzen, J. L. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14, 860–869 (2004)
Pfeil, B. E., Schlueter, J. A., Shoemaker, R. C. & Doyle, J. J. Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst. Biol. 54, 441–454 (2005)
Schlueter, J. A. et al. Mining EST databases to resolve evolutionary events in major crop species. Genome 47, 868–876 (2004)
Schlueter, J. A., Scheffler, B. E., Jackson, S. & Shoemaker, R. C. Fractionation of synteny in a genomic region containing tandemly duplicated genes across Glycine max, Medicago truncatula, and Arabidopsis thaliana . J. Hered. 99, 390–395 (2008)
Gill, N. et al. Molecular and chromosomal evidence for allopolyploidy in soybean, Glycine max (L.) Merr. Plant Physiol. 151, 1167–1174 (2009)
Bertioli, D. J. et al. An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genomics 10, 45 (2009)
Lavin, M., Herendeen, P. S. & Wojciechowski, M. F. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 54, 575–594 (2005)
Bowers, J. E., Chapman, B. A., Rong, J. & Paterson, A. H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438 (2003)
Jaillon, O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007)
Tang, H. et al. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 18, 1944–1954 (2008)
Simillion, C., Janssens, K., Sterck, L. & Van de Peer, Y. i-ADHoRe 2.0: an improved tool to detect degenerated genomic homology using genomic profiles. Bioinformatics 24, 127–128 (2008)
Krzywinski, M. et al. Circos: An information aesthetic for comparative genomics. Genome Res 19, 1639–1645 (2009)
Wang, B. B. & Brendel, V. Genomewide comparative analysis of alternative splicing in plants. Proc. Natl Acad. Sci. USA 103, 7175–7180 (2006)
Beisson, F. et al. Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol. 132, 681–697 (2003)
Fridborg, I., Kuusk, S., Moritz, T. & Sundberg, E. The Arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell 11, 1019–1032 (1999)
Barkoulas, M., Galinha, C., Grigg, S. P. & Tsiantis, M. From genes to shape: regulatory interactions in leaf development. Curr. Opin. Plant. Biol. 10, 660–666 (2007)
Lai, C. P. et al. Molecular analyses of the Arabidopsis TUBBY-like protein gene family. Plant Physiol. 134, 1586–1597 (2004)
Herve, C. et al. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 149, 1462–1477 (2009)
Stone, S. L. et al. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl Acad. Sci. USA 98, 11806–11811 (2001)
Skoneczka, J., Saghai Maroof, M. A., Shang, C. & Buss, G. R. Identification of candidate gene mutation associated with low stachyose phenotype in soybean line PI 200508. Crop Sci. 49, 247–255 (2009)
Saghai Maroof, M. A., Glover, N. M., Biyashev, R. M., Buss, G. R. & Grabau, E. A. Genetic basis of the low-phytate trait in the soybean line CX1834. Crop Sci. 49, 69–76 (2009)
Meyer, J. D. F. et al. Identification and analyses of candidate genes for Rpp4-mediated resistance to Asian soybean rust in soybean (Glycine max (L.) Merr.). Plant Physiol. 150, 295–307 (2009)
Jaffe, D. B. et al. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 13, 91–96 (2003)
Salamov, A. A. & Solovyev, V. V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10, 516–522 (2000)
Yeh, R. F., Lim, L. P. & Burge, C. B. Computational inference of homologous gene structures in the human genome. Genome Res. 11, 803–816 (2001)
Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003)
McCarthy, E. M. & McDonald, J. F. LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics 19, 362–367 (2003)
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997)
Beckstette, M., Homann, R., Giegerich, R. & Kurtz, S. Fast index based algorithms and software for matching position specific scoring matrices. BMC Bioinformatics 7, 389 (2006)
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994)
Acknowledgements
We thank N. Weeks for informatics support and C. Gunter for critical reading of the manuscript. We acknowledge funding from the National Science Foundation (DBI-0421620 to G.S.; DBI-0501877 and 082225 to S.A.J.) and the United Soybean Board.
Author Contributions Sequencing, assembly and integration: J. Schmutz, S.B.C., J. Schlueter, W.N., U.H., E.L., M.P., D. Grant, S.S., D. Goodstein, K.B., A.S., J.G. and D.R. Annotation: J.M., T.M., J.J.T., J.C., D.X., J.D., Z.T., L.Z., N.G., T.J., M.L., X.-C.Z. and G.S. EST sequencing: G.D.M., T.S., T.U., M.B., D.S., B.V., K.S. and H.T.N. Physical mapping: Y.Y., M.F.G., R.A.W. and R.C.S. Genetic mapping: D.H., J. Specht, Q.S. and P.C. Writing/coordination: S.A.J.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This Supplementary file contains Genome sequencing, assembly and mapping; Annotation results; Computational analysis of nodulation genes in the soybean genome; Soybean transcription factors and Detail of QTL trait experiments bolstered by the available genome sequence. It also includes Supplementary Figures S1-S11 with Legends, Supplementary Tables S1-S4, S6-S7 and S9 (please see separate files for Tables S5 and S8) and Supplementary References. (PDF 1637 kb)
Supplementary Table S8
This file shows computational analysis of putative nodulation genes in soybean. (DOC 163 kb)
Supplementary Table S5
This data file shows gene clusters of angiosperm and soybean. (TXT 4780 kb)
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence (http://creativecommons.org/licenses/by-nc-sa/3.0/), which permits distribution, and reproduction in any medium, provided the original author and source are credited. This licence does not permit commercial exploitation, and derivative works must be licensed under the same or similar licence.
About this article
Cite this article
Schmutz, J., Cannon, S., Schlueter, J. et al. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 (2010). https://doi.org/10.1038/nature08670
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08670
This article is cited by
-
Characterization of the pyruvate kinase gene family in soybean and identification of a putative salt responsive gene GmPK21
BMC Genomics (2024)
-
De novo genome assembly of a high-protein soybean variety HJ117
BMC Genomic Data (2024)
-
A genome-wide study of the lipoxygenase gene families in Medicago truncatula and Medicago sativa reveals that MtLOX24 participates in the methyl jasmonate response
BMC Genomics (2024)
-
Genome-wide identification, structural characterization and gene expression analysis of the WRKY transcription factor family in pea (Pisum sativum L.)
BMC Plant Biology (2024)
-
Whole-genome sequencing and comparative genomics reveal candidate genes associated with quality traits in Dioscorea alata
BMC Genomics (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.