Abstract
Tissue image analysis (tIA) is emerging as a powerful tool for quantifying biomarker expression and distribution in complex diseases and tissues. Pancreatic ductal adenocarcinoma (PDAC) develops in a highly complex and heterogeneous tissue environment and, generally, has a very poor prognosis. Early detection of PDAC is confounded by limited knowledge of the pre-neoplastic disease stages and limited methods to quantitatively assess disease heterogeneity. We sought to develop a tIA approach to assess the most common PDAC precursor lesions, pancreatic intraepithelial neoplasia (PanIN), in tissues from KrasLSL-G12D/+; Trp53LSL-R172H/+; Pdx-Cre (KPC) mice, a validated model of PDAC development. tIA profiling of training regions of PanIN and tumor microenvironment (TME) cells was utilized to guide identification of PanIN/TME tissue compartment stratification criteria. A custom CellMap algorithm implementing these criteria was applied to whole-slide images of KPC mice pancreata sections to quantify p53 and Ki-67 biomarker staining in each tissue compartment as a proof-of-concept for the algorithm platform. The algorithm robustly identified a higher percentage of p53-positive cells in PanIN lesions relative to the TME, whereas no difference was observed for Ki-67. Ki-67 expression was also quantified in a human pancreatic tissue sample available to demonstrate the translatability of the CellMap algorithm to human samples. Together, our data demonstrated the utility of CellMap to enable objective and quantitative assessments, across entire tissue sections, of PDAC precursor lesions in preclinical and clinical models of this disease to support efforts leading to novel insights into disease progression, diagnostic markers, and potential therapeutic targets.
Similar content being viewed by others
Main
Tissue image analysis (tIA) is emerging as a robust and powerful tool for objectively assessing stained tissue sections through sophisticated object recognition schemes and staining quantitation.1, 2, 3, 4 Pathologist-guided tIA uniquely enables quantification of tissue features across an entire tissue section, something that is not feasible with manual evaluation, and can be implemented in discovery-, preclinical, and clinical-stage environments.5 Furthermore, tIA captures numerous parameters, which characterize the morphometric, distribution, and biomarker staining features of each cell in significant detail—parameters that can be evaluated to develop novel insights into disease. Diseases that exist in complex tissue environments, those that require quantitative associations between multiple biomarkers to understand disease mechanisms, and those with molecular and morphometric heterogeneity pose significant challenges for manual examination and scoring of tissue. In these instances, the complexity and heterogeneity of the tissue environment can be characterized in great detail by tIA-based methods using unbiased objective tissue scoring solutions developed to complement existing approaches.6, 7
Pancreatic cancer is one such disease where tIA-based methods have the potential to complement current approaches because of the level of disease and tissue complexity.8, 9 Invasive pancreatic ductal adenocarcinoma (PDAC) is among the most lethal of all solid malignancies with a 5-year survival rate of 5%.10, 11 Early detection of localized PDAC in some cases enables surgical resection, resulting in survival rates as high as 25%, and is considered the only treatment approach with curative potential.11, 12, 13 However, only a minority (10–20%) of cases are identified early with localized tumor, and the remaining cases are characterized by invasive disease.13, 14 Advancements in chemotherapeutics and targeted therapies have provided modest improvements in survival for later-stage PDAC. However, these therapies are often confounded by highly heterogeneous tumors and survival benefits remain measured in weeks, with the majority of patients succumbing to recurrent disease.15, 16 Early detection of PDAC for more patients and the development of new therapeutics tailored to heterogeneous disease biology could have a significant impact upon patient survival, but these developments necessitate a better understanding of pathogenesis and the molecular changes that occur in the pancreas giving rise to PDAC.17
PDAC is often characterized by genetically and morphometrically heterogeneous tumor cell populations and a dynamic inflammatory infiltrate that begins during PanIN stages of the disease highlighting the uniqueness and complexity of each individual PDAC tumor.8, 9, 18 The PDAC tumor microenvironment (TME) drives inflammation, disrupts the normal tissue architecture through the deposition of fibrotic matrices, and limits the diffusion of therapeutic drugs.8, 19, 20, 21, 22 Taken together, these factors create significant challenges for researchers and clinicians to develop robust, objective, and quantitative assessments of PanIN and PDAC lesions for diagnostic and therapeutic purposes. Furthermore, assessing contextual tissue phenotypes relative to genomic and gene expression data is challenging, given the level of heterogeneity observed in PDAC and the limited tissue-based end points generated by manual evaluation of pancreatic tissue. tIA has the potential to address some of these key challenges and complement current efforts to identify novel drugs and diagnostics. Indeed, utilizing tIA in other cancer types, such as breast cancer, has provided novel insights into disease biology and biomarkers, and suggests that similar paradigms could be valuable for PDAC studies.7, 23
Pancreatic intraepithelial neoplasia (PanIN) is one of the three non-invasive forms of pancreatic neoplasia, and is the most common precursor to invasive PDAC.11, 14, 24, 25 PanIN lesions are stratified into four categories, based on the histologic appearance of the epithelium of the small caliber pancreatic ducts: PanIN-1A, PanIN-1B, PanIN-2, and PanIN-3. PanIN-3 is considered to be the direct precursor to PDAC, whereas PanIN-1 and PanIN-2 are not currently recognized to have clinical significance (reviewed in Hruban et al.26). A number of genetically engineered mouse models have been established based on mutations observed in human PDAC and can be used as disease models for developing and testing potential tIA solutions.27, 28 Specifically, the KPC mouse model is a validated and clinically relevant PDAC model that develops all stages of PanIN lesions and relies on conditionally activated Kras and p53 functions.28, 29 In the present study, we will describe a tIA-based platform for objectively quantifying biomarker expression in PanIN lesions from KPC mouse pancreata, and will discuss the potential utility of this platform to support novel PDAC studies. In addition, to extend the proof-of-concept demonstration of the tIA platform, the methods developed for KPC mice were adapted for use in human tissue and Ki-67 biomarker expression in PanIN lesions in human pancreatic tissue was evaluated.
Materials and methods
Human PDAC Tissue
IRB-approved, formalin-fixed tumor samples from pancreatic cancer patients were provided by Conversant Bio (Huntsville, AL, USA).
Animals
KrasLSL-G12D/+; Trp53LSL-R172H/+; Pdx-Cre (KPC) mice have been described previously.29 Mice were genotyped at weaning by a commercial vendor (Transnetyx, Cordova, TN, USA) and maintained under specific pathogen-free conditions according to the established institutional guidelines under the authority of a UK Home Office project license (Guidance on the Operation of Animals, Scientific Procedures Act 1986). All experimental protocols were approved by the UK Home Office. Four animals were used for the current work.
Ultrasound Analysis
PDAC tumors in KPC mice were imaged via ultrasound using the VEVO2100 high-frequency-imaging system. Mice were anesthetized using 1.5 l/min oxygen/isofluorane mixture. Vital signs were monitored on the VEVO2100 (VisualSonics, Ontario, Canada). Reference images were captured using the abdominal package in the B-mode, and three images per pancreas (10 fields/analysis) were captured via 3D-Mode combined with the B-Mode for volume analysis following the manufacturer's instructions (VisualSonics). KPC tumors are highly fibrotic in nature and were visibly distinguishable compared with a normal pancreas in the ultrasound field.
Immunohistochemistry and Histopathology
Tissue samples procured from four KPC mice and one pancreatic cancer patient were immediately placed in 10% formalin buffered with PBS. Formalin-fixed paraffin-embedded slides were processed for antigen retrieval and stained. The following antibodies were used for IHC: anti-Ki-67 (SP6) (Thermo Scientific, Waltham, MA, USA) and anti-p53 (CM5; Vector Laboratories, Burlingame, CA, USA).
Whole-Slide Scanning
Slides were digitized utilizing Aperio’s CS bright-field scanning system at Flagship Biosciences’ tissue analysis laboratory. For optimal scanning quality, 5–10 focal points were set for each individual slide, and then scanned at the × 20 scanner setting. Every image of a digitized slide was evaluated for scanning artifacts and scans were repeated if artifacts were present. In short, slide scans need to be in focus, without artifacts (such as those caused by improperly or incompletely cleaned glass slides), and free of stitching artifacts or other faults that could hamper generation of accurate data from tIA. Only scans that passed Flagship’s quality-assessment criteria were included in the study.
Manual Annotation and Image Analysis
Digital images of each tissue section were annotated to establish regions of interest (ROIs) around target tissue and to exclude staining and tissue artifacts (eg, tissue folds, artifacts in the mounting media, and so on). Pancreatic ducts displaying changes consistent with PanIN-1, -2, and -3 lesions with a diameter of <1 mm, and the immediately adjacent TME, were included as the target tissue for analysis in this study.27 Within the scope of this study, PanIN lesions were not subset to define the ROI, rather ROIs were defined by regions containing any grade of PanIN lesion and not containing PDAC or normal pancreatic tissue. tIA was performed with the CellMap software package (Flagship Biosciences, Westminster, CO, USA). Nuclei were identified using the software system based on hematoxylin staining, and biomarker staining was quantified in each nucleus based on the optical density of DAB staining. A DAB optical-density threshold was established to quantify each nucleus as either positive or negative for biomarker staining. All annotations and analysis mark-ups were assessed by a board-certified veterinary pathologist to verify annotation and algorithm performance and accuracy.
Statistical Analysis
Intra-rater reliability was assessed using intra-class correlation coefficients (ICC).30 In a fully crossed design, ICC was calculated as consistency in a two-way model, and measured the similarity between rank orders.31 Two raters, a pathologist counting cells manually and the CellMap algorithm counted total cell numbers in the two tissue compartments (tumor and TME) of interest for this study for three randomly placed scoring boxes (ie, N=6). A scheme for interpreting similar inter-rater reliability coefficients has been suggested,32 and was utilized as a guideline for interpreting the strength of the ICC. ‘Substantial’ agreement was considered for ICC>0.60 based on the guidelines proposed by Landis and Koch.
Results
Manual Annotations Established Tissue Regions for Analysis
Pancreatic tissue from KPC mice was composed of three distinct tissue compartments: (1) PanIN lesion (PanIN-1 through 3), (2) TME, and (3) normal pancreatic tissue and PDAC. The cells comprising PanIN lesions and the surrounding TME were of interest in this study and preliminary CellMap tIA algorithms were developed to attempt automated selection of only the tissue compartments of interest. For the IHC-stained tissue sections evaluated in this study, cells were detected by the algorithm based on nuclear hematoxylin staining and, therefore, nuclear staining and morphometric parameters were assessed for binning cells into each respective tissue compartment. The preliminary algorithms, however, did not adequately exclude normal pancreatic tissue, given the similarity in nuclear presentation between normal pancreatic tissue, the PanIN lesions, and the TME or efficiently exclude regions of PDAC where present. Therefore, manually establishing a ROI through annotations was necessary as a first and preliminary step in the analysis process to focus subsequent automated tissue identification and separation.
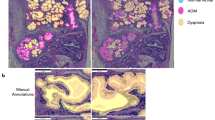
A manual annotation strategy was devised, which included all PanIN stages and TME cells while excluding staining and histology artifacts, PDAC and normal pancreatic tissue, and was applied to each digital image for analysis (Figure 1). A large inclusive annotation, which defines the ROI, was placed around each tissue section in samples where the majority of the tissue section comprised PanIN lesion and TME cells, and smaller exclusionary annotations were used to remove normal pancreatic tissue cells and artifacts from analysis (Figure 1a and b). For some tissue sections, a significant portion of the tissue contained normal pancreatic tissue (Figure 1c). Small inclusive annotations were used to capture PanIN lesion and TME cells in these cases where only small islands of target tissue within normal pancreatic tissue were present (Figure 1c and d).
Representative images of the manual annotations used in this study are shown. (a) An example of a whole-tissue section from a KPC mouse containing predominately the target tissue (PanIN and TME) for analysis is shown with the annotation mask overlaid in green. A large inclusive annotation (continuous green line) is drawn to include the target tissue, whereas smaller exclusionary annotations (dashed green line) are used to exclude non-target tissue and artifacts. (b) A higher magnification (× 10 mag.) example field from the KPC mouse tissue section in (a) is displayed. Green annotations are example exclusionary annotations around normal pancreatic tissue. (c) An example of a whole-tissue section from a KPC mouse containing predominately non-target tissue is shown. Small inclusive annotations are drawn to include only the target tissue (PanIN and TME), whereas minimal exclusionary annotations are needed to exclude non-target tissue and artifacts. (d) A higher magnification field (× 10 mag.) from the KPC mouse tissue section in (c) is displayed. Green annotations are example inclusion annotations around the target tissue. All panels are from Ki-67 IHC-stained tissue sections. The black boxes in (a and c) highlight the location of higher-magnification fields displayed in (b and d), respectively.
PanIN/TME Tissue Compartment Segmentation was Guided by tIA
Molecular changes unique to both PanIN lesions and the TME occur in pancreata during disease progression, and the separation of these distinct tissue compartments during analysis is advantageous for developing an understanding of early PDAC development. Although manual annotation allowed for the removal of normal pancreatic tissue from analysis, development of automated tIA tools was necessary to separately analyze PanIN lesions (PanIN-1 through 3, not distinguished by grade) and the surrounding TME. Manually identifying image analysis parameters (eg, nuclear area, nuclear diameter, and so on) for separating cells in PanIN lesions from the surrounding TME was particularly difficult because the nuclei of the PanIN lesions had very similar morphometric and staining features as TME cells. Therefore, the CellMap algorithm and statistical analysis methods were utilized to identify morphometric and staining differences between PanIN lesions and the TME to identify cell compartment stratification parameters (Figure 2a).
Tissue image analysis utilized to identify parameters for PanIN/TME compartment stratification scheme. (a) A schematic of the process by which tIA profiling of user-identified training PanIN and TME tissue regions can be used to identify tissue compartment stratification parameters is outlined. (b) Separate annotations are drawn around PanIN lesions (yellow annotations) in a training region of tissue and are indicated by black arrows (× 10 mag.). (c) A higher magnification (× 20 mag.) of a training PanIN lesion captured by an annotation for analysis (top panel) is shown. Each cell within the annotated region was detected by the algorithm and profiled to capture the staining and morphometric characteristics of PanIN lesion cells (bottom panel). (d) An annotation was drawn around the TME adjacent to PanIN lesions (cyan annotation) to train the algorithm to detect the TME tissue compartment (× 10 mag.). (e) A higher magnification image (× 20 mag.) of the TME region is shown (top panel), and was analyzed to profile the cells in the TME (bottom panel) adjacent to PanIN lesions. For mark-up images shown in (c and e) (bottom panels), blue-marked cells are nuclei identified by the algorithm that are negative for biomarker staining and red-marked cells are nuclei identified by the algorithm that are positive for biomarker staining. All panels are taken from p53 IHC-stained KPC mouse tissue sections. (f) Statistical methods were used to assess which of the ~100 tIA cell parameters profiled for each tissue compartment were the most differentiating between PanIN lesions (blue histogram) and the TME (red histogram). Three candidate parameters were selected as promising tissue compartment stratification features. The black bar in each plot denotes the gate for each image analysis parameter that best discriminates between PanIN lesion and TME cells; this gate was determined in a semi-automated manner using statistical tools to assess discriminating power.
Selective ROIs were drawn around representative PanIN lesion cells and TME cells as training regions of tissue. Each region for analysis was carefully placed around representative cells to ensure profiling of only the appropriate cells for each tissue compartment. Approximately one hundred parameters relating to the staining and morphometric features of nuclei within each training tissue compartment were profiled using the CellMap algorithm (Figure 2b–e). Statistical analysis of the histogram distributions of the PanIN lesions and TME cells in each training region of tissue was performed for each of the ~100 parameters profiled. Those tIA parameters identified as most distinguishing between PanIN lesion and TME cells were selected as candidates for stratification criteria (Figure 2f). Three parameters were chosen based on the statistical analysis; thresholds were defined for each parameter candidate; and these cell selection criteria were ultimately integrated into a custom CellMap algorithm to evaluate biomarker expression in PanIN lesion and TME cells as separate tissue compartments.
Automated CellMap Counting of PanIN Lesion and TME Cells was Consistent with Manual Pathologist-Based Counting
Biomarker expression positivity gates were set in the CellMap algorithm with a pathologist's guidance once the cell compartment selection criteria were integrated, and the algorithm was applied to three randomly placed boxes on one p53 IHC-stained KPC mouse tissue sample (Figure 3a). The randomly placed boxes were also evaluated by a board-certified veterinary pathologist, and the pathologist's and algorithm-counting concordance for total cell counts in the PanIN lesion and TME tissue compartments was assessed. A linear correlation was observed between pathologist-based counting and CellMap counting for both total cell counts and total p53-positive cell counts in PanIN lesions and the TME as expected (Figure 3b and c). The concordance of CellMap-based counts with pathologist counts, intra-rater reliability, was evaluated using the ICC.30 The ICC value for CellMap- vs pathologist-based total cell counts was 0.8 (95% CI: (0.12, 0.97)), indicating substantial consistency between the two sets of counts.32 The end point of interest for this proof-of-concept study, percent biomarker-positive cells, was calculated and was found to have similarly strong concordance between algorithm- and pathologist-based scores (Figure 3d). The ICC value for CellMap- vs pathologist-based percent p53+ scores was >0.95, similarly supporting the notion that the CellMap algorithm was scoring in a manner consistent with a pathologist.
Assessment of CellMap algorithm consistency with pathologist-based counting of representative tissue areas. (a) A representative counting box (green annotation box, 300 μm × 300 μm) was placed on a KPC mouse pancreas tissue sample and is shown in the left panel. The tissue sample was stained with p53 IHC. The CellMap algorithm was applied to each box, and p53-positive and -negative cells in the TME cell compartment (middle panel) and PanIN lesion cell compartment (right panel) were counted. For each mark-up image, blue-marked cells are p53-negative nuclei in the respective tissue compartment, red-marked cells are p53-positive cells in the respective tissue compartment, and green-marked cells are those cells excluded from biomarker analysis and counting (× 20 mag.). (b) Pathologist vs CellMap algorithm total cell number counts for three randomly placed counting boxes (300 μm × 300 μm) are displayed for the TME and PanIN lesion cell compartments. The dashed line represents a linear fit of the data, and is overlaid on the plot for reference. (c) Similarly, pathologist vs CellMap algorithm total p53-positive cell counts within the three randomly placed counting boxes are displayed, and a dashed line representing the linear fit of the data is shown for reference. (d) The end point measurement for this study, the percentage of p53-positive cells in each respective tissue compartment, is displayed for each of the three randomly placed counting boxes for both TME and PanIN lesion compartments and demonstrates strong agreement between algorithm-based values and pathologist-based values.
PanIN Lesion and TME p53 and Ki-67 Expressions were Assessed Across Whole-Slide Images Using tIA
The CellMap-based tIA platform developed herein was used to quantify expression of p53 and Ki-67 in PanIN lesion and TME cells across whole-tissue images of KPC mouse pancreata. Cell-by-cell analysis of biomarker expression across whole-tissue sections was a task that would have been difficult to or impossible to perform with manual assessments, while reaching an acceptable level of accuracy, given the number of cells analyzed ranging in the tens of thousands to over one hundred thousand cells. A qualitative assessment of the tissue sections indicated an increased p53 expression in PanIN lesion cells, which was consistent with the expected impact on protein expression of the p53 mutation present in KPC mice following loss of heterozygosity of the wild-type allele 33, 34 (Figure 4a). Four p53 IHC-stained KPC mouse pancreata tissue sections were evaluated using the algorithm (Figure 4b). A significant increase in the percentage of p53-positive cells in PanIN lesion cells relative to the TME cell compartment was observed, as expected, and provided a quantitative reflection of the qualitative assessment of the tissue (Figure 4c). Approximately 27 000–178 000 cells were assessed to derive the percent p53-positive cell score in each tissue compartment for each sample, which was a substantial increase in sampling of cells to determine the end point measure when compared with a typical manual assessment on the order of hundreds of cells. Ki-67 expression was similarly quantified to assess the proliferative level of PanIN lesion cells relative to surrounding TME cells (Figure 4d and e). Tissue sections from four KPC mouse pancreata were assessed using the algorithm, and the percentage of Ki-67-positive cells was found to be similar between the TME and PanIN lesion tissue compartments (Figure 4f). A low percentage of Ki-67-positive cells was observed in both tissue compartments, and was consistent with a lower proliferative index often observed in PanIN lesions.35 Similar to the assessment of p53 in KPC mice, ~21 000–150 000 cells were sampled by the algorithm in each tissue compartment across whole-slide images, and represented a robust measure of the percentage of Ki-67-positive cells.
Whole-slide image analysis of p53 and Ki-67 expression in PanIN lesion and TME cells. (a) A representative image of a p53 IHC-stained tissue section is shown. (b) The CellMap algorithm was applied to the p53 IHC-stained tissue sections to quantify p53 biomarker expression in PanIN lesion and TME cells separately. The left panel is an analysis mark-up of the TME-based analysis in a representative image field and shows predominately negative p53 staining in the TME. The right panel demonstrates the PanIN lesion-based analysis in the same image field, and shows predominately positive p53 staining in PanIN lesions. For each mark-up image, blue-marked cells are p53-negative nuclei in the respective tissue compartment, red-marked cells are p53-positive cells in the respective tissue compartment, and green-marked cells are those cells excluded from biomarker analysis and counting. (c) A summary of p53 staining in PanIN lesion and TME cells across the entire tissue sections is shown for four KPC mouse tissues analyzed, and indicates substantially more p53-positive cells in PanIN lesions. Results are presented as the percent of total cells within the respective tissue compartment positive for p53 staining, and the mean value is displayed. Error bars represent ± one s.d. around the mean value. (d) A representative image of a Ki-67 IHC-stained tissue section is shown. (e) Analysis of the TME tissue compartment is shown in the left panel, and demonstrates a mix of Ki-67-positive and -negative nuclei. Similarly, a mix of Ki-67-positive and -negative nuclei are observed in PanIN lesions and are shown in the right panel. For each mark-up image, blue-marked cells are p53-negative nuclei in the respective tissue compartment, red-marked cells are p53-positive cells in the respective tissue compartment, and green-marked cells are those cells excluded from biomarker analysis and counting. (f) A summary of Ki-67-staining analyses in PanIN lesion and TME cells for entire tissue sections from four KPC mice is shown. Similar levels of Ki-67-positive nuclei between the two tissue compartments were observed, and results are presented as the mean percentage of Ki-67-positive cells in each tissue compartment for the four samples analyzed. Error bars depict ± one s.d. around the mean value. All images in panels (a, b, d, and e) are displayed at × 20 magnification.
tIA Tools Developed for KPC Mice were Translated to Assess Human Samples
Successful application of tIA tools for the assessment of biomarker expression in PanIN lesion and TME cells from the KPC mouse model raised the question as to whether similar approaches could be used to assess human tissue. To test this possibility, the approach described herein for assessing KPC mice was utilized to develop a CellMap-based fit-for purpose algorithm solution for assessment of Ki-67 expression in a representative human tissue sample available for study as a proof-of-concept for translating the approach utilized in the preclinical mouse model to human tissues (Figure 5a). tIA profiling of representative PanIN lesion and TME training regions of tissue, and statistical analyses, were utilized to guide the selection of two tIA parameters as stratification candidates (Figure 5b). The selected tissue stratification parameter candidates, and corresponding thresholds, were integrated into a custom CellMap algorithm that resulted in robust stratification of PanIN and TME tissue compartments in the human sample (Figure 5c and d, respectively). As a proof-of-concept, the percentage of Ki-67-positive cells in the PanIN lesion and TME tissue compartments were determined using the algorithm, and approximately twice as many Ki-67-positive cells were observed in PanIN lesions relative to the TME cell compartment for the human sample analyzed (Figure 5e). Whereas this finding was inconsistent with the similar analysis of KPC mice from this study, the mild increase in Ki-67-positive cells in PanIN lesions in human tissue was consistent with previous findings.35
Application of the PanIN image analysis paradigm to a human pancreas tissue sample. (a) A representative image field displaying PanIN lesions in the human pancreatic tissue sample used in this study is shown. The tissue had been stained with Ki-67 IHC. (b) Training regions of PanIN lesion and TME cells were profiled and two cell features, one feature also selected for KPC mice and one new feature, were selected as candidates for tissue compartment stratification parameters. The histogram distributions of each parameter are shown, and the black bar denotes the cut point that optimally stratifies PanIN lesion cells from TME cells. (c) An analysis mark-up image is displayed demonstrating the performance of the CellMap algorithm selection of PanIN lesion cells using the tissue compartment stratification scheme and quantification of Ki-67 staining. (d) Displayed is an analysis mark-up image showing the performance of the algorithm to select only the TME cells and quantify Ki-67 staining in this tissue compartment. For (c and d), blue-marked cells are Ki-67-negative cells in the respective tissue compartment, red-marked cells are Ki-67-positive cells in the respective tissue compartment, and green-marked cells are those excluded from biomarker expression analysis. (e) A summary of the percentage of Ki-67-positive cells analyzed in the human tissue sample (N=1) is displayed for both the PanIN lesion and TME tissue compartments. This proof-of-concept analysis demonstrates the utility of the CellMap software for assessing biomarker expression in PanIN lesions in human pancreas tissue samples. Images in panels (a, c, and d) are shown at × 20 magnification.
Discussion
Identification and stratification of PanIN lesions and the surrounding TME can be a challenging task considering the varied histologic appearance and lesion classification scheme, but can be performed by a skilled pathologist enabled by the complexities of the human brain. Subsequent quantification of biomarker staining, however, is more difficult for a pathologist evaluating a whole-slide image, and is only made possible when a small representative area of tissue is manually counted. Interestingly, the opposite is true for tIA approaches. Stratification of PanIN lesion cells from TME cells is challenging, even with sophisticated computational approaches, which still fall short of the human brain, although quantification of biomarker staining across an entire tissue section is trivial once the correct cells are identified. The present study highlights this dichotomy, but also provides an example of the utility of tIA, which is overseen by a pathologist to guide the development of powerful analysis algorithms that perform well in PDAC.
Importantly, the tIA solutions presented in this proof-of-concept study were developed to complement the skills of a trained pathologist by providing an objective and quantitative measure of biomarker staining across entire tissue samples and across larger study cohorts. In this theme, pathology input was sought at each phase of algorithm development and refinement to ensure accurate identification of the correct cell subsets, and all analyses (eg, cell detection and biomarker scoring) were reviewed by a board-certified veterinary pathologist to verify that each tissue sample was evaluated accurately. By combining the powerful image analysis approaches described herein with pathologist oversight, we present an image analysis platform that could be utilized to advance PDAC biomarker and therapeutic development in both research and clinical environments by enabling assessments of pre-neoplastic lesions.
Separate analysis of each cell-type compartment in this study was particularly difficult because PanIN and TME cells presented with significant overlap of basic measures of nuclear architecture (eg, nuclear diameter, eccentricity, and area), making manual identification of a tissue compartment stratification scheme difficult. tIA was utilized to profile cells in user-identified training areas of PanIN lesion and TME cells, and guided the identification of a tissue compartment stratification scheme. The tissue compartment stratification scheme was integrated into the CellMap algorithm and was able to segment PanIN lesion cells from TME cells in both human and KPC mouse pancreas tissue. In addition to the challenge outlined above, further limitations of the developed algorithm include its inability to differentiate between PanIN-1, 2, and 3. In particular, this limitation is because of the algorithm being designed to separate neoplastic tumor epithelium from stroma. Digital subclassification of PanIN would require separate algorithm development. Currently, manual evaluation of ductal lesions by a pathologist is the only accepted way to perform this subclassification.
The refined CellMap algorithm resulting from this work represented a platform that could be utilized to characterize hundreds of parameters relating to biomarker expression and the morphometric features of individual cells in the PanIN and TME cell compartments, data which could be further analyzed to elucidate novel mechanisms or molecular interactions in PanIN and PDAC biology. tIA-based approaches, such as the CellMap platform described herein, can enable these novel insights by generating a diverse data set describing each unique tumor in great detail, and similar paradigms in breast cancer have identified new prognostic factors related to TME cells and have improved prognostic assessments by genomic profiling.7, 9, 23
Quantification of biomarker expression in each cell across an entire tissue section was relatively straightforward once the central challenge of correctly stratifying the PanIN lesion and TME tissue compartments was solved. As a proof-of-concept of the CellMap tIA platform, Ki-67 and p53 expression were assessed in KPC mouse pancreata. tIA scoring of each sample provided robust quantitative values for p53 and Ki-67 expression in the KPC mouse model because each value was based on sampling tens of thousands, to over one hundred thousand, cells in each tissue compartment for each sample. Sampling cells across an entire tissue section is particularly advantageous for PanIN and PDAC, where high degrees of tissue heterogeneity are expected and manual scoring of representative areas is likely to inadequately capture the key features of the tissue. The CellMap tIA platform was able to capture the range of heterogeneity on a cell-by-cell basis across an entire tissue section, and these data provide the level of dimensionality likely needed to identify complex relationships between tissue phenotypes, genomic profiling data, and molecular changes during disease progression from non-neoplastic to PanIN to PDAC.
References
Potts SJ, Johnson TD, Voelker FA et al, Multiplexed measurement of proteins in tissue in a clinical environment. Appl Immunohistochem Mol Morphol 2011; 19: 494–498.
Potts SJ, Krueger JS, Landis ND et al, Evaluating tumor heterogeneity in immunohistochemistry-stained breast cancer tissue. Lab Invest 2012; 92: 1342–1357.
Angell HK, Gray N, Womack C et al, Digital pattern recognition-based image analysis quantifies immune infiltrates in distinct tissue regions of colorectal cancer and identifies a metastatic phenotype. Br J Cancer 2013; 109: 1618–1624.
Mofidi R, Walsh R, Ridgway PF et al, Objective measurement of breast cancer oestrogen receptor status through digital image analysis. Eur J Surg Oncol 2003; 29: 20–24.
Aeffner F, Wilson K, Bolon B et al, Commentary: roles for pathologists in a high-throughput image analysis team. Toxicol Pathol 2016; 44: 825–834.
Shah SP, Morin RD, Khattra J et al, Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 2009; 461: 809–813.
Yuan Y, Failmezger H, Rueda OM et al, Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med 2012; 4: 157ra43.
Clark CE, Hingorani SR, Mick R et al, Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res 2007; 67: 9518–9527.
Samuel N, Hudson TJ . The molecular and cellular heterogeneity of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol 2012; 9: 77–87.
Siegel R, Naishadham D, Jemal A . Cancer statistics, 2013. CA Cancer J Clin 2013; 63: 11–30.
Hernandez YG, Lucas AL . MicroRNA in pancreatic ductal adenocarcinoma and its precursor lesions. World J Gastrointest Oncol 2016; 8: 18–29.
Cameron JL, Crist DW, Sitzmann JV et al, Factors influencing survival after pancreaticoduodenectomy for pancreatic cancer. Am J Surg 1991; 161: 120–124; discussion 4–5.
National Cancer Institute (NCI). SEER Cancer Statistics Review (CSR) 1975-2011, 2012. Available at: http://seer.cancer.gov/archive/csr/1975_2011/browse_csr.php?sectionSEL=3&pageSEL=sect_03_table.08.html.
Guo J, Xie K, Zheng S . Molecular biomarkers of pancreatic intraepithelial neoplasia and their implications in early diagnosis and therapeutic intervention of pancreatic cancer. Int J Biol Sci. 2016; 12: 292–301.
Conroy T, Desseigne F, Ychou M et al, FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. New Eng J Med 2011; 364: 1817–1825.
Burris HA 3rd, Moore MJ, Andersen J et al, Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol 1997; 15: 2403–2413.
Sharma MR, Schilsky RL . Role of randomized phase III trials in an era of effective targeted therapies. Nat Rev Clin Oncol 2012; 9: 208–214.
Makohon-Moore A, Brosnan JA, Iacobuzio-Donahue CA . Pancreatic cancer genomics: insights and opportunities for clinical translation. Genome Med 2013; 5: 26.
Feig C, Gopinathan A, Neesse A et al, The pancreas cancer microenvironment. Clin Cancer Res 2012; 18: 4266–4276.
Bachem MG, Zhou S, Buck K et al, Pancreatic stellate cells—role in pancreas cancer. Langenbecks Arch Surg 2008; 393: 891–900.
Chang DZ, Ma Y, Ji B et al, Mast cells in tumor microenvironment promotes the in vivo growth of pancreatic ductal adenocarcinoma. Clin Cancer Res 2011; 17: 7015–7023.
Provenzano PP, Cuevas C, Chang AE et al, Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell 2012; 21: 418–429.
Beck AH, Sangoi AR, Leung S et al, Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med 2011; 3: 108ra13.
Hruban RH, Adsay NV, Albores-Saavedra J et al, Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol 2001; 25: 579–586.
Maitra A, Fukushima N, Takaori K et al, Precursors to invasive pancreatic cancer. Adv Anat Pathol 2005; 12: 81–91.
Hruban RH, Maitra A, Goggins M . Update on pancreatic intraepithelial neoplasia. Int J Cin Exp Pathol 2008; 1: 306–316.
Hruban RH, Adsay NV, Albores-Saavedra J et al, Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res 2006; 66: 95–106.
Gopinathan A, Morton JP, Jodrell DI et al, GEMMs as preclinical models for testing pancreatic cancer therapies. Dis Model Mech 2015; 8: 1185–1200.
Hingorani SR, Wang L, Multani AS et al, Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005; 7: 469–483.
Shrout PE, Fleiss JL . Intraclass correlations: uses in assessing rater reliability. Psychol Bull 1979; 86: 420–428.
Hallgren KA . Computing inter-rater reliability for observational data: an overview and tutorial. Tutor Quant Methods Psychol 2012; 8: 23–34.
Landis JR, Koch GG . The measurement of observer agreement for categorical data. Biometrics 1977; 33: 159–174.
Lane DP, Benchimol S . p53: oncogene or anti-oncogene? Genes Dev 1990; 4: 1–8.
Hezel AF, Kimmelman AC, Stanger BZ et al, Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 2006; 20: 1218–1249.
Klein WM, Hruban RH, Klein-Szanto AJ et al, Direct correlation between proliferative activity and dysplasia in pancreatic intraepithelial neoplasia (PanIN): additional evidence for a recently proposed model of progression. Mod Pathol 2002; 15: 441–447.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
FA, NTM, MP, JCB, JM, JK, and GDY were full-time employees of Flagship Biosciences Inc. during the course of data collection and creation of this manuscript. MOP was a full-time employee of Gilead Sciences during the course of data collection and creation of this manuscript. The remaining author declares no conflict of interest.
Rights and permissions
About this article
Cite this article
Aeffner, F., Martin, N., Peljto, M. et al. Quantitative assessment of pancreatic cancer precursor lesions in IHC-stained tissue with a tissue image analysis platform. Lab Invest 96, 1327–1336 (2016). https://doi.org/10.1038/labinvest.2016.111
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2016.111