Abstract
Conventional histopathology with hematoxylin & eosin (H&E) has been the gold standard for histopathological diagnosis of a wide range of diseases. However, it is not performed in vivo and requires thin tissue sections obtained after tissue biopsy, which carries risk, particularly in the central nervous system. Here we describe the development of an alternative, multicolored way to visualize tissue in real-time through the use of coherent Raman imaging (CRI), without the use of dyes. CRI relies on intrinsic chemical contrast based on vibrational properties of molecules and intrinsic optical sectioning by nonlinear excitation. We demonstrate that multicolor images originating from CH2 and CH3 vibrations of lipids and protein, as well as two-photon absorption of hemoglobin, can be obtained with subcellular resolution from fresh tissue. These stain-free histopathological images show resolutions similar to those obtained by conventional techniques, but do not require tissue fixation, sectioning or staining of the tissue analyzed.
Similar content being viewed by others
Main
With rare exceptions, histopathological techniques cannot be used for in situ diagnosis as they require the tissue to be frozen or fixed, thinly sliced and stained with devitalizing dyes before the tissue can be observed under a microscope. A real-time, in situ technique for acquiring histopathological images would thus be tremendously advantageous in situations where the tissue removal imposes risk and delay required for fixation, sectioning and staining. Ideally, in situ stain-free histopathology could be used in the setting of tumor surgery to examine the margins of a resection cavity for residual neoplastic tissue.
Coherent Raman imaging (CRI), including coherent anti-Stokes Raman scattering (CARS) microscopy1, 2 and stimulated Raman scattering (SRS) microscopy,3, 4, 5, 6 allows chemical imaging based on intrinsic vibrational properties of the molecules in the tissue and thus does not require staining or labeling. The more recent SRS microscopy techniques offer further advantages: (1) elimination of image artifacts due to nonresonant background, (2) excitation spectra identical to well-documented spontaneous Raman spectra, (3) a linear relationship between signal and concentration of target molecules. Compared with spontaneous Raman scattering, signal levels are amplified by orders of magnitude by virtue of stimulated excitation of molecular vibrations, allowing video-rate image acquisition speeds.7, 8 Owing to nonlinear excitation, CRI enables intrinsically three-dimensional sectioning1, 4, 9 and does not require physical sectioning of the tissue. Efficient signal detection in reflection of thick samples,8, 10 as well as biocompatibility of laser excitation intensities11, 12 have been demonstrated, allowing stain-free, in vivo imaging in mice7, 8, 13, 14, 15, 16 and humans.8, 17
The properties of CRI have inspired us18 and others19, 20, 21 to image diseases with CARS microscopy and development of miniaturized clinical instrumentation is underway.22, 23, 24, 25 This work describes the development of multicolor SRS imaging with contrast originating from lipids, protein and red blood cells, aiming to resemble the most widely used stain in histopathology, hematoxylin and eosin (H&E). We demonstrate ex vivo that the key diagnostic features appreciated with H&E staining are well visualized with CRI in tissue from wild-type mice, as well as mouse models of various diseases of the central nervous system.
MATERIALS AND METHODS
Imaging Setup and Processing
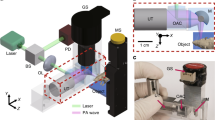
CRI requires two synchronized pulse trains to achieve stimulated excitation of the vibrational transition. In narrowband SRS, the frequency bandwidth of such lasers is chosen to be smaller than the typical linewidth of Raman transition and the difference in frequency of the two center frequencies can be tuned to excite a single vibration at a time. Compared with multiplex excitation of multiple vibrations simultaneously using broadband lasers, this approach maximizes imaging speed. We use a passively mode-locked Nd:YVO4 laser (PicoTrain, High Q Laser) at 1064 nm (7 ps pulse-duration, 76 MHz repetition rate) to provide the first beam, known as the Stokes beam. A portion of this beam is frequency-doubled and used to pump an optical parametric oscillator (Levante Emerald, APE GmbH) to provide the second tunable beam (650–1000 nm tuning range), known as the pump beam. The two pulse trains are then overlapped in space and time, aligned into a laser scanning microscope (FV300, Olympus) and focused into the sample with a high numerical aperture (NA) water-immersion lens (UPlanSApo × 60, Olympus).2, 4
For SRS microscopy we measure the intensity loss of the pump beam due the excitation of molecular vibrations in the focus. Sensitive detection of the signal requires the implementation of a high-frequency modulation transfer scheme to distinguish the SRS signal from linear sample absorption or laser intensity fluctuation. This relies on the fact that SRS can only occur if both pump and Stokes are incident on the sample. By modulating the Stokes beam at a known frequency with an acousto-optic modulator and measuring the modulation transfer to the originally unmodulated pump beam at the same frequency, SRS can be detected specifically. If the modulation frequency is faster than the typical laser noise (eg 10 MHz), high sensitivity detection is achieved at the moderate laser power required for medical imaging. Experimentally, the transmitted light from the sample is collected with a high NA oil condenser (Nikon), filtered to block the modulated Stokes beam (CARS890/220, Chroma) and detected with a large-area photodiode (FDS1010, Thorlabs). The modulation transfer is measured by a lock-in amplifier (SR844, Stanford Research), whose signal is fed into the microscope to provide the signal for a pixel. All images were sampled with 512 × 512 pixels at a ∼30 μs pixel dwell time, which is limited by the speed of the lock-in amplifier. Recently we have developed improved detection electronics and achieved imaging speeds up to video-rate (30 frames per second).4, 8
For the multicolor imaging, three individual scans per region in the tissue are acquired with one beam fixed at 1064 nm and the other at 816.7 nm (CH2-stretching vibration), 810.5 nm (CH3-stretching vibration) and 700 nm (hemoglobin two-color two-photon absorption (TPA)). Fast tuning between these three wavelengths can be achieved within a few seconds by changing the Lyot filter and the cavity length of the optical parametric oscillator. Images are acquired with the microscope scanning software and processed in ImageJ. Pseudo-colors can be assigned arbitrarily, for example to mimic H&E-stained sections. In other cases we use a red–green–blue (RGB) look-up table. The green and red channels show CH2 and hemoglobin images, respectively. The blue channel is a CH3-CH2 difference image thresholded to show only nuclei (see Results section). To normalize for the curvature of the field of view we calculated a featureless reference image by averaging and smoothing about one hundred SRS images acquired the same day from different regions and dividing the CH2 and CH3 images before subtraction. Linear look-up tables are applied for the RGB coloring. H&E-like colors were applied using the ‘multiply layers’ blend mode in Adobe Photoshop.
Animal Handling and Preparation
All animals were treated in compliance with Harvard IACUC protocols #29-01 and #10-02. Mice were euthanized using CO2 or by an overdose of ketamine and xylazine, followed by cervical spine dislocation. Organs were harvested and imaged immediately. We used a mouse brain matrix (Harvard Apparatus) to produce 1 mm thick tissue slices and mounted them stably for imaging between a No.1 coverslip and a coverslide using a ∼480 μm spacer (Grace Bio-Labs). For the comparison with H&E-stained micrographs, the brain was coronally sectioned and one-half was immediately fixed in 10% formalin for standard histopathological evaluation, while the other was imaged without further preparation as described above. Comparison is shown from mirror images.
Invasive glioma models
We created primary brain glioma models from immunocompromised mice. The human glioma stem cell lines were derived from Brigham and Women’s Hospital patients undergoing surgery according to approved protocols. These xenografts were derived from glioblastoma biopsy samples and implanted. Dissected xenografts were washed in artificial cerebrospinal fluid and manually dissociated to single cells. Red blood cells were removed using Lympholyte-M (Cedarlane). The cells were cultured in DMEM/F12 (with L-glutamine, Invitrogen) medium containing glucose (0.3%), penicillin/streptomycin (50 μg/ml), Apo-transferrin (0.1 mg/ml), Progesterone (20 nM), Sodium selenite (30 nM), putrescine (60 μM), insulin (25 μM/ml), sodium bicarbonate (3 mM), HEPES (10 mM), 20 ng/ml EGF and 20 ng/ml FGF. Live cells were counted using a hemocytometer and Trypan blue exclusion. The Brigham and Women’s Hospital IACUC protocol number is 000995.
Metastasis model
We created breast cancer metastasis models from nude mice (Harlan Laboratory). MDA-MB-231 breast cancer cells were cultured in Eagle’s Minimum Essential Medium (ATCC, containing nonessential amino acids, 2 mM L-glutamine, 1 mM sodium pyruvate, and 1500 mg/l sodium bicarbonate) supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and grown in a 5% CO2 incubator at 37 °C. Metastatic breast cancer were established by intracranially injecting 1 × 105 MDA-MB-231 cells (in 2 μl PBS) at 1 mm posterior to bregma, lateral 2 mm and 2.5 mm depth from skull surface. The Brigham and Women’s Hospital IACUC protocol number is 04662.
Stroke model
We used a transient model of focal brain ischemia in C57/BL6J mice (Charles River lab). Under an operating microscope (Leica), the origin of right common carotid artery (CCA) was ligated with 6-0 silk suture. A loose ligature and a microvascular clip were placed on the CCA before the origin of the internal carotid artery. An incision was made between the two sutures through which the silicone coated tip (HeraeusKulzer) of a 7-0 filament was advanced to the origin of the middle cerebral artery (MCA). With a flexible fiber probe fixed to the right skull above the MCA, regional cerebral blood flow was monitored with laser Doppler flowmetry (PF2B, Perimed) to confirm successful MCA occlusion and reperfusion. The filament was withdrawn 30 min later. Mice were euthanized at day 3 post-ischemia. The Massachusetts General Hospital IACUC protocol number is 2007N000168.
Demyelination model
We created relapsing-remitting EAE (R-EAE) from C57Bl/6 mice (Charles-River). Each mouse was immunized subcutaneously (into the flanks) with 100 μg of MOG35-55 (MEVGWYRSPFSRVVHLYRNGK), 400 μg of Mycobacterium tuberculosis (strain H37Ra; Difco) in complete Freund’s adjuvant (CFA, Difco). One hundred nanogram of Bordetella pertussis toxin (List Biological Laboratories via Cedarlane) was given intravenously at the time of immunization and 2 days later. The Brigham and Women’s Hospital IACUC protocol number is 04576.
RESULTS
Mechanism of Multicolor Image Contrast
Different molecules in the sample can be selectively imaged by tuning the difference frequency of the excitation beams into the target vibrational resonance (see Methods). Figure 1a shows the vibrational spectra of the three major components of cells and tissue (lipids, protein and water) with characteristic peaks for the CH2 and CH3-stretching vibrations at 2845 cm−1 and 2940 cm−1, respectively. While imaging of DNA and RNA has recently been demonstrated with CRI in the finger-print region of Raman spectra,26 the above three species are present in higher concentration and allow for faster imaging speed, as desirable for medical in vivo imaging.18
Stain-free histologic imaging with multicolor coherent Raman imaging. (a) Vibrational spectra of the major constituents of tissue: lipids, protein and water. Arrows indicate Raman shifts at which imaging is performed. (b–d) stimulated Raman scattering (SRS) images of a live C2C12 mammalian cell acquired at the CH2-stretching vibration at 2845 cm−1 (b) and CH3-stretching vibration at 2940 cm−1 (c). Multicolor image (d) generated from images (b, c) with the green channel (CH2 image) showing the cell-body and the blue channel (thresholded CH3-CH2 difference image) highlighting the nuclear morphology including a bright nucleolus. (e–h) SRS images of fresh ex vivo brain tissue acquired at CH2-stretching vibration at 2845 cm−1 (e), CH3-stretching vibration at 2940 cm−1 (f), and vibrationally off-resonant showing two-color two-photon absorption of hemoglobin at a sum frequency of 23 700 cm−1 (g). Multicolor image (h) generated from images (e–g) with the green channel (CH2 image) highlighting cytoplasm and myelin sheaths, blue channel (thresholded CH3-CH2 difference image) showing the nuclear morphology, and the red channel (hemoglobin image) highlighting the red blood cells. (i) Hematoxylin & eosin (H&E)-stained micrograph from the same region in the brain. (j) Same multicolor image as (h) with a different pseudo-color scheme, chosen to mimic the appearance of an H&E-stained micrograph, illustrates the similar image content and appearance of stain-free images and H&E-stained sections. Scale bar, 25 μm.
Figures 1b–d show SRS images of a C2C12 mammalian cell at these two bands. Consistent with spontaneous Raman spectroscopy,27 the CH2 image (Figure 1b) mainly highlights lipids in the cytoplasm, while lipid-poor nuclei appear dark and featureless. The CH3 image (Figure 1c) shows signal from both lipids and proteins (Figure 1a). The same cytoplasmic features can be seen as in the CH2 image, but there is additional detail from within nuclei. We have demonstrated that a quantitative protein image,17 which shows a similar distribution of proteins in both the nucleus and cytoplasm, can be extracted from the CH2 and CH3 images based on the Raman ratios in Figure 1a. Here, we aim to generate a vibrational counterstain to the CH2 image that accentuates nuclei similarly to hematoxylin component of an H&E stain. To do so, we subtract the CH2 from the CH3 image such that the cytoplasmic signal essentially vanishes and then threshold the resulting difference image to only show nuclear signal. This is possible in a robust way because of the low CH2 signal inside the nucleus. Figure 1d illustrates the resulting multicolor overlay showing the cell-body information from the CH2 image in green and nuclear information from a thresholded CH3-CH2 difference image in blue. Diagnostically important subnuclear features such as nucleoli are visible in the thresholded CH3-CH2 difference image, demonstrating that it is a true nuclear counterstain rather than merely the inverse of the CH2 image.
Figures 1e–j illustrate similar contrast obtained in fresh mouse brain tissue. Figure 1e and the green component of Figure 1h show the CH2 signal from lipid-rich myelinated axons28 and unmyelinated neuropil. Additionally, the vibrationally off-resonant image (Figure 1g) has contrast due to TPA9, 16, 29 and shows individual red blood cells within capillaries due to hemoglobin absorption. Overlaying these three images results in a single three-color, stain-free histological image shown in Figure 1h. As the choice of the color scheme is arbitrary, it is possible to choose the look-up tables to mimic an H&E-stained section (Figure 1j). This allows easy comparison with traditional histopathological techniques (Figure 1i). Comparison of Figures 1i and j reveals that the stain-free image is free of the vacuolization observed in the H&E-stained section because the fresh tissue imaging eliminates the need for freezing, fixation or sections and hence associated tissue processing artifacts.
Tissue Imaging of Various Organs
To further demonstrate the universal utility of this multicolor contrast we imaged a variety of organs of a wild-type mouse (Figure 2). The myocardium image (Figure 2b) shows myocytes with numerous lipid droplets, central nuclei and a rich vascular network. The typical morphology of the renal glomerulus inside Bowman’s capsule with tufts of capillaries and surrounding renal tubules is seen in the image of the kidney (Figure 2c). Variable lipid storage within hepatocytes is seen in the liver (Figure 2d). In the lung, the vast capillary network of the alveolar walls is highlighted in the red channel (Figure 2e). Characteristic cross striations and capillaries around the myocytes are easily identified in the skeletal muscle image (Figure 2f). Clusters of germinal cells and larger blood vessels are seen in the ovary (Figure 2g). Figure 2h shows two different layers of mouse skin including the stratum corneum layer (top) with a cobblestone arrangement of cells and a protein-rich hair shaft in blue. A deeper image of the epidermis (bottom) collected simply by changing the level of focus highlights many nuclei in the stratum basale. The germinal centers with abundant green cytoplasm surrounded by mature lymphocytes with higher nuclear-to-cytoplasmic ratio, as well as scattered red blood cells, are observed in the white pulp of the spleen (Figure 2i).
Multicolor stain-free images of various mouse organs (green: CH2 image; blue: CH3-CH2 difference image; red: hemoglobin image) of (a) brain, (b) heart, (c) kidney, (d) liver, (e) lung, (f) muscle, (g) ovary, (h) skin with stratum corneum (top) and stratum basale (bottom) and (i) spleen. Scale bar, 25 μm.
Tissue Imaging of Normal Mouse Nervous System
Imaging of gray matter (Figure 3a) demonstrates neuronal cell bodies in blue, reflecting a strong signal in the CH3-CH2 channel within in a background of the lipid-rich neuropil. Linear processes in green, due to high CH2 signal, are axons (Figures 3a–c). The white matter from the corpus callosum is dominated by densely packed axons with interspersed oligodendrocytes (Figure 3h). The cytoarchitecture of various regions including the thalamus (b), the dentate gyrus (c) and the granular cell layer of the cerebellar cortex (g) is easily distinguished, as compared with the mirror images stained with Luxol-H&E (Figures 3d–f)
Multicolor stain-free images of various brain regions in a wild-type mouse in comparison with paraffin-embeded, hematoxylin & eosin (H&E) and luxol-stained sections. (green: CH2 image; blue: CH3-CH2 difference image; red: hemoglobin image) of (a) cortex, (b) thalamus, (c) dentate gyrus, (g) cerebellum, and (h) corpus callosum. (d–f, i, j) show H&E/luxol-stained section of corresponding regions.
Diagnostic Features in Brain Lesions
In addition to demonstrating image quality and histopathological information in healthy tissue, we imaged mouse models of four relatively common brain pathologies including glioblastoma, metastases, demyelination and stroke. We aimed at recognizing tumor from normal tissue, tumor margins, inflammatory infiltration and evidence of demyelination and cell death. For this reason, we imaged mouse models of invasive, high-grade glioma derived from human brain-tumor stem cells, and breast cancer metastasis, as well as demyelination and stroke models. Fresh tissue samples were bisected to acquire corresponding stain-free and paraffin-embedded, H&E-stained images from the same region of the same animal (Figure 4).
Comparison of multicolor, label-free images with H&E-stained sections of primary mouse models of glioma and metastatic breast cancer. Images display practically identical diagnostic features as visualized with stimulated Raman scattering (SRS). (a, b, e, f) Two-color SRS images (green: CH2 image; blue: CH3-CH2 difference image) and (c, d, g, h) hematoxylin & eosin-stained micrographs from the same region of the same animal. (a–d) are acquired from mouse models of primary brain tumor and (e–h) from models of breast cancer brain metastasis. Hypercellularity is noticeable in both primary tumor and metastasis. The primary brain-tumor morphology (a, c) shows residual interspersed axons (highlighted in green), while metastasis (e, g) lacks this feature due to their epithelioid, cohesive and less infiltrative character. Primary and secondary tumors can be further distinguished at the tumor margin, where primary tumor cells invade the healthy tissue (b, d) while metastases produce a sharp margin at the interphase of tumor and normal tissue (f, h). Scale bar, 25 μm.
In comparison with the healthy brain, increased cellularity, a fundamental histopathological feature of neoplasia, is made obvious in SRS and H&E images for both primary brain tumors (Figures 4a–d) and brain metastases (Figures 4e–h). The remaining myelinated axons interspersed between tumor cells are visible in the center of the infiltrating glioma (Figure 4a) in contrast with the solid, expansile metastatic tumor (Figure 4e). Images of tumor margins illustrate the diffusely invasive nature of glial cells along white matter tracts of the corpus callosum (Figure 4b), whereas a sharply defined margin is seen between metastasis and adjacent normal brain tissue (Figure 4f). A three-dimensional image stack of the tumor margin further highlights this critical diagnostic feature (Supplementary Video S1).
Among other pathologies that can mimic brain cancer on MRI and CT are tumefactive demyelination and subacute stroke.30 Figure 5 and Supplementary Videos S2 and S3 show multicolor SRS images of mouse models of experimental autoimmune EAE and ischemic stroke. The comparison with corresponding H&E images shows similar diagnostic features. Lipid-laden macrophages and perivascular lymphocytes (Figures 5a and c) were present in areas of demyelination. Tissue vacuolization due to cerebral edema was seen as black holes (Figures 5b and d), which correlated well with the presence of clear vacuoles on H&E-stained sections. Foamy macrophages and condensation of nuclei correlated with apoptosis.
Stimulated Raman scattering (SRS) label-free images of demyelination and stroke mouse models. (a) Two-color SRS image (green: CH2 image; blue: CH3-CH2 difference image) of a demyelinating lesion in a mouse with experimental allergic encephalomyelitis (EAE). Perivascular inflammatory cells and histiocytes with large lipid droplets from phagocytized myelin are visible. Red blood cells in a capillary are highlighted by arrows (hemoglobin image, red, not used in this picture). (b) Two-color SRS image of brain tissue in a mouse stroke model 3 days post stroke. Macrophages with many small lipid droplets are dispersed throughout the tissue. Dark areas represent edema fluid. (c, d) Hematoxylin & eosin-stained micrographs of (c) EAE and (d) stroke model from the same region. Scale bar, 25 μm.
DISCUSSION
We report the successful acquisition of microscopic, multicolored images of diagnostic quality on various fresh tissues, without the need for tissue fixation, processing or staining. We acquired images of multiple normal mouse organs and compared them with routine histopathology. Recognition of various tissue types of different organs was easy and characteristic histological features were demonstrated with a clarity that approaches that of traditional H&E staining. In addition, all major histologic features demonstrated in H&E-stained sections of cerebral cortex, white matter, diencephalon and cerebellum were visualized with CRI. Given the great variety of histopathological features in various conditions, we also imaged multiple regions of the normal mouse brain, as well as models of glioma, metastasis, stroke and demyelination.
The goal of this study was to demonstrate medical utility of CRI stain-free histology using a research type microscope in ex vivo tissue to motivate the design of a clinical prototype instrument in the future. We used the original instrumentation of SRS microscopy;4 the imaging speed was limited by the speed of the commercial lock-in amplifier and signal was collected in sample transmission. Since completion of the data acquisition for this paper, we developed faster detection electronics and an improved signal collection scheme, which enable SRS at video-rate and in reflection from thick tissue samples, and demonstrated SRS in vivo imaging.8 For this study we also used temporal multiplexing to acquire the individual color channels sequentially. Development of real-time multicolor imaging is underway17, 31 and will be available for the clinical prototype. TPA can also be probed at 827nm, so more robust and inexpensive fiber lasers with a limited tuning range over the CH-region of Raman spectra can be used in future. As imaging depth in CRS is limited to about 150 μm, the most immediate applications are in imaging exposed tissue such as ex vivo pathological samples, skin and in vivo intraoperative margins. The development of a fiber-coupled hand-held scanner for intraoperative use is underway.22, 23, 24, 25 Imaging of hollow organs inside the living patient will require further development of endoscopic instrumentation in the future.
Other label-free microscopy techniques used for in situ stain-free histopathology such as infrared (IR) absorption microscopy32 allow multicolor imaging; however, the spatial resolution and penetration depth into water-rich samples is limited due to the long wavelength used in IR microscopy.21 Alternative high-resolution techniques include confocal reflection, optical coherence tomography (OCT), two-photon excited auto-fluorescence (TPAF), second harmonic generation (SHG) and third harmonic generation (THG) microscopy.33, 34, 35, 36, 37 They either lack the capability for multicolor, chemical imaging (CF, OCT and THG) or have contrast limited to a few, specific molecules (TPAF and SHG).38, 39, 40 The latter techniques are more similar to specific functional stains (eg antibody stains) in traditional histopathology, which can be used after abnormalities have been recognized on H&E sections. TPAF has also been demonstrated to allow identification of nuclear and cytosolic regions41, 42 because of the absence of TPAF signal inside the nucleus, analogous to the dark nuclei seen in the CH2 CRI scans. Multicolor CRI can additionally provide a ‘counterstain’ image with positive nuclear contrast for diagnostically important subnuclear features, which makes multicolor CRI images an ideal foundation for versatile in situ histopathological tissue diagnosis.
In conclusion, we have demonstrated the use of CRI for microscopic tissue imaging in a label-free manner. Moreover, CRI can be used to generate a degree of contrast, based on the intrinsic vibrational spectrum of the molecular components of biologic tissues that exceeds other label-free optical imaging methods. Our data indicates that CRI can be used for generating high quality histological images without the need of routine tissue processing. Additional work will be required to comprehensively compile an atlas of CRI images of normal tissue and various disorders. However, a preliminary sampling of key neurologic diseases supports the diagnostic capability of CRI. These findings reinforce the possibility that CRI can become an invaluable clinical tool for intraoperative distinction of tumor from normal tissue and immediate margin assessment of in situ tissue before surgical closure.
References
Zumbusch A, Holtom GR, Xie XS . Three-dimensional vibrational imaging by coherent anti-Stokes Raman scattering. Phys Rev Lett 1999;82:4142–4145.
Evans CL, Xie XS . Coherent anti-Stokes Raman scattering microscopy: chemical imaging for biology and medicine. Annu Rev Anal Chem 2008;1:27.
Ploetz E, Laimgruber S, Berner S et al. Femtosecond stimulated Raman microscopy. Appl Phys B 2007;87:389–393.
Freudiger CW, Min W, Saar BG et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 2008;322:1857–1861.
Ozeki Y, Dake F, Kajiyama S et al. Analysis and experimental assessment of the sensitivity of stimulated Raman scattering microscopy. Optics Express 2009;17:3651–3658.
Nandakumar P, Kovalev A, Volkmer A . Vibrational imaging based on stimulated Raman scattering microscopy. New J Phys 2009;11, (article: 033026); http://iopscience.iop.org/1367-2630/11/3/033026/pdf/1367-2630_11_3_033026.pdf.
Evans CL, Potma EO, Puoris'haag M et al. Chemical imaging of tissue in vivo with video-rate coherent anti-Stokes Raman scattering microscopy. Proc Natl Acad Sci U S A 2005;102:16807–16812.
Saar BG, Freudiger CW, Reichman J et al. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science 2010;330:1368–1370.
Fu D, Ye T, Matthews TE et al. Two-color, two-photon, and excited-state absorption microscopy. J Biomed Opt 2007;12:54004–54009.
Cheng JX, Volkmer A, Book LD et al. An epi-detected coherent anti-Stokes raman scattering (E-CARS) microscope with high spectral resolution and high sensitivity. J Phys Chem B 2001;105:1277–1280.
Nan XL, Potma EO, Xie XS . Nonperturbative chemical imaging of organelle transport in living cells with coherent anti-Stokes Raman scattering microscopy. Biophysical J 2006;91:728–735.
Fu Y, Wang HF, Shi RY et al. Characterization of photodamage in coherent anti-Stokes Raman scattering microscopy. Optics Express 2006;14:3942–3951.
Fu Y, Wang HF, Huff TB et al. Coherent anti-Stokes Raman scattering imaging of myelin degradation reveals a calcium-dependent pathway in lyso-PtdCho-induced demyelination. J Neurosci Res 2007;85:2870–2881.
Fu Y, Huff TB, Wang HW et al. Ex vivo and in vivo imaging of myelin fibers in mouse brain by coherent anti-Stokes Raman scattering microscopy. Optics Express 2008;16:19396–19409.
Henry FP, Côté D, Randolph MA et al. Real-Time in vivo assessment of the nerve microenvironment with coherent anti-Stokes Raman scattering microscopy. Plastic and Reconstructive Surgery 2009;123:123s–130s.
Fu D, Ye T, Matthews TE et al. High-resolution in vivo imaging of blood vessels without labeling. Opt Lett 2007;32:2641–2643.
Fu D, Lu FK, Zhang X et al. Quantitative chemical imaging with multiplex stimulated Raman scattering microscopy. J Am Chem Soc 2012;134:3623–3626.
Evans CL, Xu X, Kesari S et al. Chemically-selective imaging of brain structures with CARS microscopy. Optics Express 2007;15:12076–12087.
Begin S, Belanger E, Laffray S et al. In vivo optical monitoring of tissue pathologies and diseases with vibrational contrast. J Biophotonics 2009;2:632–642.
Chowdary PD, Jiang Z, Chaney EJ et al. Molecular histopathology by spectrally reconstructed nonlinear interferometric vibrational imaging. Cancer Res 2010;70:9562–9569.
Meyer T, Bergner N, Bielecki C et al. Nonlinear microscopy, infrared, and Raman microspectroscopy for brain tumor analysis (Journal Paper). J Biomed Opt 2011;16:021113.
Légaré F, Evans CL, Ganikhanov F et al. Towards CARS endoscopy. Opt Express 2006;14:4427–4432.
Murugkar S, Smith B, Srivastava P et al. Miniaturized multimodal CARS microscope based on MEMS scanning and a single laser source. Optics Express 2010;18:23796–23804.
Balu M, Liu G, Chen Z et al. Fiber delivered probe for efficient CARS imaging of tissues. Optics Express 2010;18:2380–2388.
Saar BG, Johnston RS, Freudiger CW et al. Coherent Raman scanning fiber endoscopy. Optics Letters 2011;36:2396–2398.
Zhang X, Roeffaers MB, Basu S et al. Label-free live-cell imaging of nucleic acids using stimulated Raman scattering microscopy. ChemPhysChem 2012;13:1054–1059.
Slipchenko MN, Le TT, Chen HT et al. High-speed vibrational imaging and spectral analysis of lipid bodies by compound Raman microscopy. J Phys Chem B 2009;113:7681–7686.
Wang HF, Fu Y, Zickmund P et al. Coherent anti-stokes Raman scattering imaging of axonal myelin in live spinal tissues. Biophysical J 2005;89:581–591.
Matthews TE, Piletic IR, Selim MA et al. Pump-probe imaging differentiates melanoma from melanocytic nevi. Science Translational Medicine 2011;3:71ra15.
Omuro AMP, Leite CC, Mokhtari K et al. Pitfalls in the diagnosis of brain tumours. Lancet Neurology 2006;5:937–948.
Laffray S, Pagès S, Dufour H et al. Adaptive movement compensation for in vivo imaging of fast cellular dynamics within a moving tissue. PloS One 2011;6:e19928.
Gremlich HU, Yan B . Infrared and Raman Spectroscopy of Biological Materials. New York: Marcel Dekker, Inc., Vol. 24 2001.
Master BR, So PTC . Biomedical Nonlinear Optical Microscopy. New York: Oxford University Press, Inc., 2008.
König K, Ehlers A, Riemann I et al. Clinical two-photon microendoscopy. Microsc Res Techn 2007;70:398–402.
Tearney GJ, Brezinski ME, Bouma BE et al. In vivo endoscopic optical biopsy with optical coherence tomography. Science 1997;276:2037–2039.
Tai S, Tsai TH, Lee WJ et al. Optical biopsy of fixed human skin with backward-collected optical harmonics signals. Optics Express 2005;13:8231–8242.
Witte S, Negrean A, Lodder JC et al. Label-free live brain imaging and targeted patching with third-harmonic generation microscopy. Proc Natl Acad of Sci 2011;108:5970.
Zipfel WR, Williams RM, Christie R et al. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc Natl Acad Sci U S A 2003;100:7075–7080.
Li CQ, Pastila RK, Pitsillides C et al. Imaging leukocyte trafficking in vivo with two-photon-excited endogenous tryptophan fluorescence. Optics Express 2010;18:988–999.
Campagnola PJ, Millard AC, Terasaki M et al. Three-dimensional high-resolution second-harmonic generation imaging of endogenous structural proteins in biological tissues. Biophysical J 2002;82:493–508.
Palero JA, de Bruijn HS, van der Ploeg van den Heuvel A et al. Spectrally resolved multiphoton imaging of in vivo and excised mouse skin tissues. Biophysical J 2007;93:992–1007.
König K . Clinical multiphoton tomography. J Biophotonics 2008;1:13–23.
Acknowledgements
We thank Dr John Sims for the mouse stroke model, Chrissy Rooney for the invasive glioma model, Dr Fa-Ke Lu and Srinjan Basu for the cell images, and Dr Anita Huettner for early pathology work. Funding: CWF and BGS thank Boehringer Ingelheim Fonds for a Ph.D. Fellowship and the Army Research Office for a National Defense Science and Engineering Graduate Fellowship, respectively. LO acknowledges the Italian Society of Multiple Sclerosis (FISM, Cod 2008/B/5). PLD is a Harry Weaver neuroscience scholar of the National Multiple Sclerosis society and CW acknowledges NIH (R01NS049263). The work of X Xu was supported by the NSF award 0958345. This work was supported by the NIH Director’s Pioneer Award (to XSX) and NIH/NIBIB T-R01 (grant 1R01EB010244-01).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Harvard University has filed a patent applications based on coherent Raman imaging instrumentation.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
Conventional histopathology with hematoxylin & eosin has been the gold standard for histopathology. An alternative multicolored method to visualize tissue in real-time using coherent Raman imaging has been developed. The histopathological images show resolution similar to conventional techniques but do not require tissue fixation, sectioning or staining of the tissue analyzed.
Rights and permissions
About this article
Cite this article
Freudiger, C., Pfannl, R., Orringer, D. et al. Multicolored stain-free histopathology with coherent Raman imaging. Lab Invest 92, 1492–1502 (2012). https://doi.org/10.1038/labinvest.2012.109
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2012.109
Keywords
This article is cited by
-
Development and prospective validation of an artificial intelligence-based smartphone app for rapid intraoperative pituitary adenoma identification
Communications Medicine (2024)
-
Instant diagnosis of gastroscopic biopsy via deep-learned single-shot femtosecond stimulated Raman histology
Nature Communications (2022)
-
Vibrational imaging for label-free cancer diagnosis and classification
La Rivista del Nuovo Cimento (2022)
-
Stimulated Raman histology facilitates accurate diagnosis in neurosurgical patients: a one-to-one noninferiority study
Journal of Neuro-Oncology (2022)
-
Detection of glioma infiltration at the tumor margin using quantitative stimulated Raman scattering histology
Scientific Reports (2021)