Abstract
Species tree methods have provided improvements for estimating species relationships and the timing of diversification in recent radiations by allowing for gene tree discordance. Although gene tree discordance is often observed, most discordance is attributed to incomplete lineage sorting rather than other biological phenomena, and the causes of discordance are rarely investigated. We use species trees from multi-locus data to estimate the species relationships, evolutionary history and timing of diversification among Australian Gehyra—a group renowned for taxonomic uncertainty and showing a large degree of gene tree discordance. We find support for a recent Asian origin and two major clades: a tropically adapted clade and an arid adapted clade, with some exceptions, but no support for allopatric speciation driven by chromosomal rearrangement in the group. Bayesian concordance analysis revealed high gene tree discordance and comparisons of Robinson–Foulds distances showed that discordance between gene trees was significantly higher than that generated by topological uncertainty within each gene. Analysis of gene tree discordance and incomplete taxon sampling revealed that gene tree discordance was high whether terminal taxon or gene sampling was maximized, indicating discordance is due to biological processes, which may be important in contributing to gene tree discordance in many recently diversified organisms.
Similar content being viewed by others
Introduction
One of the difficulties in the inference of species trees from multiple gene trees is overcoming situations in which individual gene trees differ from one another, a situation that poses significant challenges for traditional methods of combining information from multiple loci via concatenation (Edwards et al., 2007; Kubatko and Degnan, 2007; Huang et al., 2010; Salichos and Rokas, 2013). Discordance between gene trees can be caused by both stochastic (for example, incorrect gene tree estimation) and technical (for example, paralogous sequences) errors (Chung and Ané, 2011). A number of biological processes, such as incomplete lineage sorting (ILS) and gene flow are known to create further discordance between gene trees (Maddison, 1997) and the underlying species tree. Species tree methods represent a conceptual shift in phylogenetics in that the estimation gene and species trees is considered separately. These methods aim to account for discordance between gene trees in the estimation of species trees but make different inferences regarding the source of the discordance (Chung and Ané, 2011).
In addition to accounting for gene tree discordance, the advent of fossil-calibrated phylogenies utilizing multiple genes and individuals for each species can significantly increase power to test for the association of linking biogeographic events with the diversification history of species (Drummond and Rambaut, 2007; McCormack et al., 2010), although discordance between gene trees can have an adverse effect on the ability to estimate rates of divergence and, thus, divergence dates (Burbrink and Pyron, 2011). It is due to this ability to deal with certain levels of gene tree discordance that makes species tree methods also particularly useful for reconstructing the evolutionary history of recent and rapid radiations that have historically been problematic to reconstruct using more traditional phylogenetic methods (Edwards et al., 2007; McCormack et al., 2010; Rowe et al., 2010; Salichos and Rokas, 2013). However, species tree methods largely attribute discordance to ILS rather than technical errors or other biological phenomena (Chung and Ané, 2011). Despite some studies investigating the impact of discordance on the accuracy and interpretation of phylogenetic analyses (for example, Leaché, 2009; Reid et al., 2012), few empirical studies attempt to investigate the degree of discordance present or its potential sources.
The focal organismal group for our study—Gehyra—is a large genus of geckos from the family Gekkonidae (Han et al., 2004; Russell and Bauer, 2002), comprising 36 species occupying a wide range of habitats from Indochina throughout most of Oceania and Melanesia (King, 1979; Russell and Bauer, 2002). The Australian Gehyra radiation represents the bulk of the group’s diversity comprising 19 largely endemic species (Horner, 2005; Sistrom et al., 2009). The Australian Gehyra radiation has proven to be taxonomically troublesome in the past, as considerable genetic, karyotypic and allozyme variation does not manifest in easily recognizable morphological variation. Thus, many species comprise multiple morphological isolates, distinct chromosome races, allozymeoperational taxonomic units and mitochondrial clades (King, 1979, 1982, 1983, 1984; Moritz, 1984, 1986, 1992; Sistrom et al., 2009; Sites and Moritz, 1987). As a relatively recent radiation (King, 1984), gene tree discordance in Gehyra is expected owing to ILS (Edwards, 2009). However, gene tree discordance in recent radiations can also be caused by low locus signal resulting in multiple optimal topologies being supported by a single gene. Furthermore, there is evidence of prevalent gene flow between species in the Gehyra variegata species complex (Sistrom et al., 2013) that may contribute to gene tree discordance. To evaluate the causes of gene tree discordance in the Australian Gehyra radiation, we conducted a Bayesian concordance analysis (BCA) to examine discordance varying both individual and gene sampling. If observed gene tree discordance is due to a technical error (for example, incorrect assignment of individuals to species or the sequencing of a paralogous locus), high discordance with maximal locus sampling and minimal individual sampling and reduced discordance with maximal individual sampling and minimal locus sampling would be expected. Alternatively, if discordance is due to biological processes, changing sampling efforts should have limited effects on observed patterns of discordance. In addition, we calculate Robinson–Foulds distances (RFDs) to determine whether discordance is attributable to uncertainty within loci or due to discordance in topologies recovered from different loci. If uncertainty is high within loci, low levels of phylogenetic signal may explain overall discordance; however, if uncertainty is high between loci, biological processes are more likely to be the cause of observed discordance.
Despite the lack of satisfactory taxonomic resolution, hypotheses regarding the evolutionary history of Gehyra have been considered to plausibly account for the history of the group, even when lacking substantial empirical justification. King (1979, 1983, 1984) summarized many of these assumptions, including (1) A recent Asian origin for Gehyra (King, 1984); (2) Australian Gehyra form two major species complexes (Mitchell, 1965)—the G. variegata complex characterized by small bodied species associated with arid regions (King, 1979) and the Gehyra australis species complex characterized by larger bodied animals associated with tropical, subtropical and monsoonal regions (King, 1983); and (3) that radiation within these two complexes was due to allopatric divergence and chromosomal rearrangement with sequential radiations of allopatrically derived species from 2n=44 chromosome ancestor(s), to 2n=42 chromosome species and to 2n=40 and 2n=38 chromosome species simultaneously. However, King’s proposal was criticized as premature given the incomplete taxonomy of the genus and lack of data demonstrating reproductive isolation of allopatric chromosome races (Moritz, 1992; Sites and Moritz, 1987). Using a combination of species tree reconstruction, molecular dating methods and ancestral state reconstruction, we evaluate these hypotheses regarding the evolution of the Australian Gehyra radiation. We test the validity of the hypothesis that Australian Gehyra result from a single, recent colonization event from a Melanesian ancestor that subsequently split into a large-bodied, tropically adapted australis species group and a small-bodied, arid-adapted variegata species group. We also test whether the King (1984) model of diversification driven by chromosomal rearrangement in allopatric populations is supported by our species tree approach.
Our phylogenetic framework allows us to evaluate support for the various hypotheses regarding this historically difficult group and also explore the sources of uncertainty in our species tree reconstructions and associated interpretation. We discuss our results and the importance of exploring the source of gene tree discordance.
Methods
Sampling
All tissue samples were obtained from Australian museum collections (Australian Biological Tissue Collection at the South Australian Museum, Western Australian Museum) or sequences were available on GenBank (Supplementary Appendices S1 and S2—GenBank accession numbers will be added upon acceptance). In order to be sure of correct assignment of Gehyra species, we included a sample collected from as close as possible to the type locality, which had a corresponding museum specimen that was visually verified as being representative of the type specimen. DNA was extracted using a Puregene DNA Isolation Tissue Kit D-7000a (Gentra Systems, Minneapolis, MN, USA) following the manufacturer’s guidelines. Standard PCR methods were used to amplify the coding region of the mitochondrial gene NADH dehydrogenase subunit 2 (ND2), portions of the nuclear coding genes recombination-activating gene 1 (RAG1), prolactin receptor (PRL-R), melanocortin 1 receptor (MC1R), the first and second intron of the histone cluster 3 gene along with the contained exon region (H3) and two anonymous nuclear loci (A1 and A2). Anonymous loci were developed from the analysis of DNA fragments generated from a partial shotgun library using the GS-FLX 454 sequencing (Roche, Basel, Switzerland), isolated using the methods described in Bertozzi et al. (2012). A summary of primers used is provided in Table 1. PCR products were sequenced using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit and an ABI 3730 automated sequencer (Life Technologies, Carlsbad, CA, USA). Sequences were edited by eye and aligned at first using the Muscle plug-in in Geneious v5.3.1 (Biomatters, Auckland, New Zealand) (Drummond et al., 2010; Edgar, 2004) then refined by eye.
Estimation of rates of evolution within Gehyra
Divergence times between representatives of major Gehyra lineages were estimated from the RAG1 data due to the availability of data on Genbank and previous studies with which to compare our estimated divergence dates (Gamble et al., 2010; Sanders et al., 2008) (Supplementary Appendix S1) using Bayesian inference implemented in BEAST v1.6.1 (Drummond and Rambaut, 2010). All non-Gehyra sequences were obtained from Genbank. Monophyly of the Gekkotans in relation to other squamates is well established (for example, Gamble et al., 2010, Oliver and Sanders, 2009) and was thus assumed a priori. Model selection was determined using the Akaike Information Criterion (AIC) carried out using jModeltest v0.1.1 (Posada, 2008). A Yule branching process with a uniform prior was adopted. A relaxed clock was used and rate variation across adjacent branches was assumed to be uncorrelated. Model parameter values between taxa partitions were unlinked and the analysis run for 50 million generations, with the first 15 million discarded as burn in and every 1000th tree sampled thereafter. Output was evaluated using TRACER v1.4.1 (Drummond and Rambaut, 2010) to confirm acceptable mixing, stationarity of the MCMC parameter sampling and adequate effective sample sizes (>500). Due to the lack of Gekkotan fossils, which can be placed with enough phylogenetic precision to act as molecular clock calibrations (Sanders et al., 2008; Oliver and Sanders, 2009; Gamble et al., 2010), a number of robust external fossil calibrations were used. Our chosen calibrations are similar to those of Sanders et al. (2008) and are summarized in Table 2. All calibrations were treated as being uncertain and given lognormal distributions, in order to reflect known bias in the fossil record (Sanders and Lee, 2007). A liberal, uniform prior of 160–250 mya was placed on the base of the tree to prevent the analysis becoming stuck in an unrealistic parameter space (Drummond et al. 2006). To ensure adequate searching of the parameter space, the analysis was repeated 10 times. The posterior set of trees was summarized using TreeAnnotator (Drummond and Rambaut, 2010) before being visualized using FigTree v1.4.0 (Rambaut, 2009).
Species tree reconstruction and divergence estimation within Australian Gehyra
Sampling for the reconstruction of species relationships was based on a total of 123 individuals and the seven genes listed above. Taxon sampling included five individuals where possible from all described Gehyra species, and selected representatives of Melanesian Gehyra (G. baliola, G. barea, G. membranacruralis, G mutilata and G. oceanica) and six recently discovered species (using mtDNA screening, morphological analysis and species boundary assessment—Sistrom et al., in revision) to determine the phylogenetic placement of the Australian Gehyra in relation to Melanesian taxa. We undertook locus sampling in a hierarchical manner, sequencing a larger number of individuals for faster evolving loci (for example, ND2) compared with markers traditionally used to resolve deeper phylogenetic relationships (for example, RAG1) (see Supplementary Appendix S2 for details on the scheme for locus sampling for each individual). Attempts were made to sequence at least one individual per species for each locus; however, where this was not achieved, data were coded as missing in the *BEAST input file. Although this approach considerably increases the MCMC sampling required to reach convergence in Bayesian analysis and thus computational expense, it allows a sequence to be placed anywhere in the tree and thus is the most conservative approach to dealing with missing information from a species. Collecting sequence data in this manner is expected to have a minimal impact on analytical power (Wiens and Morrill, 2011) while reducing sequencing cost. We used a conservative approach in estimating the rate of sequence evolution by placing a normally distributed prior on the substitution rate of the RAG1 data set (see above), taken from the 95% confidence interval (CI) for rate estimation along each branch among the Gehyra in the dating analysis.
When sequences were obtained from more than one individual for a species, gametic phase of was estimated for alignments of each species using the program PHASE (Stephens et al., 2001) under default settings. Haploytpes estimated with a confidence of >90% were retained. Ambiguous sites in alleles that could not be estimated with acceptable confidence were coded as missing data. Bayesian estimation of the species level phylogeny was undertaken using *BEAST (Heled and Drummond, 2010). *BEAST utilizes a single step approach to simultaneously estimate gene trees from individual sequence alignments and the overall species tree simultaneously. Substitution models for individual genes were determined using the AIC carried out using jModeltest v0.1.1 (Posada, 2008; see Table 1), and all related parameters were estimated in *BEAST. A Yule branching process with a uniform prior was adopted, and a relaxed clock was used. Rate variation across adjacent branches was assumed to be uncorrelated for all gene trees. The mutation rate for the RAG1 gene tree was given a lognormal prior distribution with upper and lower rates representing the fastest and slowest rates observed in the broader dating analysis as represented by the 95% CIs of all branches within Gehyra in that analysis and the mean representing the average of all observed rates within Gehyra and estimated rates for all other genes. Coding loci were partitioned by codon. Model parameter values were unlinked and the analysis run for 500 million generations, with the first 50 million discarded as burn in and every 10 000th tree sampled thereafter. Output was evaluated using TRACER v1.4.1 (Drummond and Rambaut, 2010) as for the higher-level analysis. To ensure adequate searching of the parameter space, the analysis was repeated 10 times. A maximum clade credibility species tree was produced by combining the trees remaining after burn in from all runs using LogCombiner (Drummond and Rambaut, 2010) and summarized using TreeAnnotator (Drummond and Rambaut, 2010) before being visualized using FigTree v1.4.0 (Rambaut, 2009).
Gene tree discordance analysis
As gene trees inferred from different loci are often incongruent (Chung and Ané, 2011; Cranston, 2010; Degnan and Rosenberg, 2009), which can impact the statistical support of the overall estimated species tree and thus the robustness of hypotheses inferred from it, it is important to investigate the level of potential discordance between gene trees and evaluate the potential sources of this discordance. As an initial examination of discordance, individual gene trees from each of the 10 *BEAST runs were combined with LogCombiner (Drummond and Rambaut, 2010) and summarized using TreeAnnotator (Drummond and Rambaut, 2010), once 25% of the trees had been removed as burn in. Tree files were visualized using FigTree v1.4.0 (Rambaut, 2009) (Supplementary Appendix S3).
Like other species tree approaches (for example, STEM, BEST, MDC), *BEAST accounts for potential discordance between trees by attributing the discordance between trees to ILS (Larget et al., 2010). Consequently, if discordance is a result of gene flow, the method may incorrectly produce a smaller distance between lineages than expected under the coalescent model (Liu and Yu, 2011). This is of particular concern in Gehyra where admixture between species cannot be ruled out. In order to investigate the role of potential sources of gene tree incongruence, a BCA of unphased loci was used to estimate gene tree discordance (Larget et al., 2010) without making assumptions with regard to the source of that discordance. Methods for measuring gene tree discordance are still in development and require congruent sampling of individuals and species across loci (Cranston, 2010). In order to meet this requirement, we used a hierarchical approach to test our data. As the RAG1 gene tree has the most minimal sampling, all other gene trees were trimmed to match RAG1 taxon sampling (n=30). At the next level, A7, A8 and MC1R had similar sampling, so all gene trees excluding RAG1 were trimmed to have identical sampling (n=44). Finally, as ND2, H3 and PRL-R all had near complete individual sampling, as a final step in our hierarchical approach, these were trimmed to have identical sampling (n=76). Models were determined using the AIC implemented in jModeltest v0.1.1 (Posada, 2008), and all model parameters were unlinked. For each tier, individual gene trees were estimated using MrBayes v3.1 (Ronquist and Huelsenbeck, 2003). Each analysis was run for 15 million generations sampled every 1000 generations. Using the program mbsum (Larget et al., 2010), tree files from the two chains for each Bayesian analysis were combined once the first 10% of trees had been discarded as burn in. Once combined, BUCKy v1.4.0 (Larget et al., 2010) was used to conduct BCA analysis. Each BCA analysis comprised two independent runs with four chains each for two million generations sampled every 100 generations. The primary concordance tree for each BCA analysis was visualized using FigTree v1.4.0 (Rambaut, 2009), with the concordance factor (CF) for each node displayed on the tree. If incorrect assignment of individuals to species (a significant possibility with Gehyra geckos) is a cause of discordance, increasing individual sampling would decrease CFs. Conversely, if sampling of paralogous sequences was the source of error, we expected that increasing the sampling of loci would reduce CFs; however, it should be noted that ILS may generate a similar result.
To evaluate discordance caused by uncertainty in topological estimation within gene trees, we generated 95% credible tree files from the individual gene trees estimated using MrBayes and measured RFDs both within each 95% credible file and between each file using the treedist component of the PHYLIP package (Felsenstein, 2005). These comparisons were conducted in a hierarchical manner identical to the BCA analysis described above. We then normalized RFDs by the maximum RFD for each tree, presenting each distance as a percentage of contradictory splits in each tree set (Table 3). In order to test whether within-gene RFDs were significantly different from between-gene RFDs, we compared each set of within-gene RFDs to its respective between-gene set of RFDs for each comparison made using a nonparametric Kruskal–Wallis test (Kruskal and Wallis, 1952) implemented in the base package of R (R Core development Team, 2011).
In order to evaluate the potential role of selection in generating gene tree discordance, we conducted a relative rates ratio test for each locus using CRANN v1.04 (Creevey and McInerney, 2003) using the method described in Creevey and McInerney (2002). As CRANN requires uniform sampling of individuals in both the phylogeny it uses to calculate Dn and Ds values, the individual gene tree generated for each locus using MrBayes was used for these calculations. We report in Table 4 the percentage of pairwise calculations between branches on each of these trees for which selection was significant.
Ancestral state reconstruction
In order to evaluate the likely ancestral state of the Australian Gehyra, we reconstructed the ancestral state of both body form and chromosome number. This was conducted using the re-rooting method implemented in the Phytools package (Revell, 2012) in the R statistical environment (R Core Development Team, 2011). For body form, each species in the tree estimated from *BEAST was coded as either tropical or arid, for chromosome each species was coded as a chromosome number. For species where the karyotype was unknown, equal probability for each of the four chromosome numbers was assumed and the ancestral state reconstruction was used to estimate the likelihood of each state at the tip. Each analysis was conducted using a symmetrical maximum likelihood model.
Results
Estimation of rates of evolution in Gehyra
The results of the analysis of rate estimation using the RAG1 data set and a Bayesian uncorrelated relaxed clock with five external fossil calibrations (Table 2) are presented in Figure 1. Divergence dates across squamates and geckos were largely concordant with previous studies (Gamble et al., 2008a, 2010; Sanders et al., 2008). This indicates that date estimates for splits within Gehyra are likely to be reasonable given the available calibrations. Divergence of G. oceanica from G. australis and G. variegata had a point estimate of 29.74 mya (95% CI 45. 05–17.22 mya), and divergence between G. variegata and G. australis had a point estimate of 11.24 mya (95% CI 21.32–3.95 mya). From this analysis, we used the average branch rate of evolution of 0.0007 mutations per year (95% CI 0.0002–0.0019) for further species tree analyses.
Dating analysis using fossil calibrations from Table 2. Node bars represent the 95% confidence interval (CI) of divergence dates in years, and node labels represent posterior probabilities. Calibrated node bars are shown in black. Gehyra are shown to be a monophyletic member of the subfamily Gekkoninae; the split between Australian Gehyra and G. oceanica is shown to have occurred approximately 29.74 ma. (95% CI 45. 05–17.22 ma.).
Species tree reconstruction and divergence estimation within Australian Gehyra
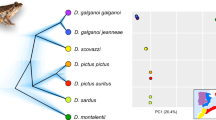
The results of species tree estimation are presented in Figure 2. Overall, posterior probabilities across the species tree appear relatively low, and BCA results confirm a high degree of discordance in the data. This could indicate uncertainty in the observed species tree and suggests that interpretations be undertaken with caution. However, as support values for species tree analyses are expected to be lower than when traditional concatenation approaches are used (Edwards, 2009) and our subsequent analyses of discordance indicate that gene tree discordance is relatively high in our data set, we believe that the recovered phylogeny represents the most likely topology for the group according to the data at hand. The species tree analyses (that is, *BEAST and BCA) find a basal split of Australian Gehyra into two clades, but the content of the two groups differs from those proposed by King. Two species, G. occidentalis and G. xenopus, that were regarded as members of the australis group by King fall in with members of his variegata group. In addition, one Melanesian species, G. membranacruralis, branches at the base of our australis group rather than with the other Melanesian species (G. oceanica, G. baliola and G. barea). A comparison of the divergence estimates for the basal splits within our revised G. variegata and G. australis (excluding G. membranacruralis) clades revealed near identical estimates: G. variegata—6.8 mya (95% CI 17.8–1.9 mya)—and G. australis—7.0 mya (95% CI 18.0–1.9 mya)—with broad overlap of the estimates of splits within each clade (Figure 2).
Gene tree discordance analysis
A visual inspection of individual gene trees from the *BEAST analysis reveals considerable discordance between genes (Supplementary Appendix S3). Analysis of hierarchically trimmed gene trees showed CFs (a measure of the percentage of gene trees which support a particular node) were low overall, indicating a high level of gene tree discordance (Figure 3). The deeper relationships between taxa in the BCA analysis at different sampling levels are considerably variable—further supporting high levels of gene tree discordance. However, the topology of the species tree attained using BCA and containing all genes shows a high degree of similarity with the *BEAST species tree reconstruction. The topologies of these trees support the basal position of the Melanesian species relative to the Australian species groups and the New Guinean G. membranacruralis, reciprocal monophyly of the G. australis and G. variegata clades and species membership of each. Average RFDs measured within and between gene trees are presented in Table 3. Distances within-gene trees were consistently lower than between-gene trees—a trend which was statistically supported by Kruskal–Wallis tests showing that all between-gene tree distances were significantly different (P>0.001) to all respective within-gene trees (Supplementary Appendix S4). This result indicates that discordance between gene trees is significantly higher than uncertainty within gene trees, suggesting that the discordance observed in the data set is more attributable to different genes displaying distinct histories than low overall power to estimate species relationships within each gene.
Results of BUCKy species tree estimation and Bayesian concordance analysis. (a) Sampling of 30 individuals and 7 genes. (b) Sampling of 44 individuals and 6 genes. (c) Sampling of 76 individuals and 3 genes. Terminal labels represent Gehyra species, and node labels represent concordance factors (CFs)—a measure of the number of gene trees across the sample that support a node. CFs were generally low regardless of whether gene or taxon sampling were maximized.
Results for the relative rates ratio test for selection are reported as the percentage of pairwise calculations between branches for each locus for which significant selection was detected. This percentage ranged from 0 to 7.61% in the nuclear loci but was 20.51% of comparisons in the mitochondrial ND2 locus.
Ancestral state reconstruction
Results from ancestral state reconstruction analyses are displayed in Figure 4. We were able to support the ancestral node of the tree as being of tropical body form (likelihood=98.0%) and the ancestral node of the Australian Gehyra as also being of tropical origin, albeit with lower support (likelihood=83.6%). Although we are able to confidently rule out 2n=38 as the ancestral karyotype of Gehyra, the remaining three states have similar likelihood at the ancestral node (2n=40–37%, 2n=42–39%, 2n=44–24%) and the node at the base of the Australian clade (2n=40–37%, 2n=42–40%, 2n=44–22%). We are therefore unable to support the King hypothesis of chromosomal evolution in Gehyra.
Discussion
Consistent topologies were recovered from both rate estimation and species tree analyses for independent runs. The rate estimation analysis places Gehyra as a monophyletic group within the subfamily Gekkoninae and both phylogenies show that the Australian Gehyra species are a largely monophyletic clade, nested within a broadly distributed assemblage of Melanesian Gehyra species. Support values among Australian Gehyra were low for the species tree analysis, despite consistent recovery of the same species tree. BCA results indicate that support values remain low whether taxon or locus sampling is maximized, indicating that the low support is likely due to gene tree discordance generated by biological processes rather than technical errors. However, due to the incomplete nature of the data matrix, there are certain circumstances in which our BCA analysis may produce misleading results (for example, if the best-sampled gene is paralogous and all others are not, CFs will not increase with addition of more genes). Additionally, if the initial individuals in Figure 3a have a higher likelihood of being misidentified, then CF may decrease with the addition of more individuals—however, as we biased our data collection towards typotypic individuals, this is unlikely. Investigation of RFDs indicates that discordance is significantly higher between gene trees rather than within, suggesting that the source of discordance is differences between genes rather than due to low or conflicting signal within genes.
The exception to monophyly of the Australian Gehyra is G. membranacruralis, a phenotypically Melanesian species from southern New Guinea, which is nested within the Australian Gehyra clade. However, this relationship is weakly supported (pp=0.45) and possibly that due to G. membranacruralis being represented by a single individual with a long branch separating it from the other sampled species, resulting in an incorrect placement of the species. It seems likely that G. membranacruralis would be more appropriately considered a close relative of the Australian radiation, an assertion that is supported by the BCA analysis, albeit also weakly (CF=0.29).
Regardless of the precise branching position of G. membranacruralis, the basal split separating the Australian clade from the Melanesian assemblage occurred between the mid-Eocene and the early Miocene and between the G. australis and G. variegata clades dates between the early Miocene and the mid Pliocene. A number of studies of Australian herpetofauna, which have diversified over similar timescales to Gehyra, have revealed some contiguous patterns with those observed in this study. For example, the shift from a mesic phenotypic state to an arid phenotypic state as seen in this study is observed over very similar timescales to Heteronotia binoei in the same biogeographic region, although H. binoei shows multiple independent origins for the arid phenotype throughout the species complex, in contrast with Gehyra (Fujita et al., 2010). The patterns observed in Gehyra contrast those observed in Rynchoedura in which diversification was observed to follow drainage basins (Pepper et al., 2011)—a distribution not mirrored by Gehyra. It appears that the diversification of Australia’s herpetofauna over the period from the late Miocene is particularly complex.
The impact of gene tree discordance
Despite the large number of samples that we used for species tree estimation, posterior probabilities of tree nodes are low overall, as were CFs in our BCA (Figures 2 and 3). The hierarchical approach to sampling we have undertaken in our BCA analysis shows that CFs remain low regardless of whether taxon or gene sampling is maximized—indicating that discordance between gene trees is the likely source of uncertainty in our data set rather than a technical error, such as misidentification of samples. In addition, RFDs show that discordance is due to differences between the topologies inferred by different genes rather than uncertainty of the topology inferred from a given gene (Table 3 and Supplementary Appendices S3 and S4). This strongly suggests that the discordance, and hence low support values in our species tree estimation, are due to biological processes rather than technical errors. However, discordance within genes revealed by RFDs (Table 3) may well be useful in determining the likely level of signal in respective loci and provide useful context in marker selection for future studies. In addition, for genes that were present in all three hierarchical RFD analyses, a slight decrease in the level of discordance is observed as individual sampling is increased, suggesting that the inclusion of more than one individual per species is increasing the phylogenetic resolution of individual genes—an expected result in recently diverged groups of organisms (Maddison and Knowles, 2006). Investigation of the role of selection in our data revealed relatively low levels, or no selection acting on the six nuclear loci (Table 4); however, the mtDNA locus—ND2 has significant selection present in 20.51% of branch comparisons—suggesting at selective processes may well be causing this locus to deviate from the true species tree. As ND2 is arguably the most widely used phylogenetic marker utilized in our study and has often been used in single locus studies, this potentially has significant implications for the results of these studies and highlights in an empirical sense the potential pitfalls of phylogenies estimated from single loci.
Discrepancies in the history of individual gene trees can result due to a number of biological processes—for example, selection, failure to coalesce following recent divergence (Knowles and Carstens, 2007), gene flow (Liu and Pearl, 2007), gene duplication (Kubatko et al., 2009) and recombination (Lanier and Knowles, 2012). In the case of Australian Gehyra, the combination of a relatively recent evolutionary history and likely ongoing diversification suggests that discordance due to ILS may be present, and complex patterns of gene flow between distinct species has been documented (Sistrom et al., 2012), indicating that horizontal gene transfer is also likely to generate discordance in the Australian Gehyra radiation. In addition, selection acting on the mtDNA locus ND2 may be causing deviation of the gene tree estimated from it, and potentially explaining discrepancies in the multi-locus species tree estimated here, and single locus studies utilizing this gene in the past (Sistrom et al., 2009; Oliver et al., 2010).
As *BEAST (and other species tree estimation methods) assumes all discordance arises from ILS (Larget et al., 2010), and as gene flow is a potential cause of discordance, it is possible that the distance between species are incorrectly assumed to be shorter than they truly are. For this reason, our substitution rates are deliberately conservative, and thus the error bars surrounding nodes in the species tree are more likely to encompass the true divergence times of species than a more restrictive prior. Distinguishing between ILS and gene flow is a significant hurdle in the estimation of species trees, and the determination of evolutionary relationship between species and development of methods to distinguish between these two processes is ongoing (Chung and Ané, 2011). Thus, despite the fact that we recovered the same topology from multiple runs with different seeds, the low support values in our species tree estimation represent genuine uncertainty in estimating the species relationships among Australian Gehyra stemming from biological processes that make estimating accurate phylogenies inherently challenging.
Thus despite allowing for more accurate reconstruction of species relationships, species tree methods can also uncover inherent uncertainty in empirical data sets that are potentially masked by traditional (that is, concatenation) approaches. Although the discovery of gene tree discordance might lower the support for phylogenetic inferences and it is sometimes considered preferential to exclude discordant loci from phylogenetic analysis (Townsend, 2007), investigating discordance can highlight biological processes affecting radiations of organisms. As the current study shows, such investigation can shed light on the evolutionary processes shaping the diversification of species.
Hypothesis 1—recent Asian origin of the Australian Gehyra
Our analyses support previous evidence (Sistrom et al., 2009; Oliver et al., 2010) that the Australian Gehyra radiation is monophyletic and derived in relation to Melanesian Gehyra. The estimated time of divergence of the Australian clade from the rest of the Melanesian assemblage covers a wide interval from the mid-Eocene to the early Miocene. This makes attributing a particular biogeographic event to the introduction of Gehyra to Australia difficult; however, it does coincide with the collision of the Australian tectonic plate with the Ontong Java plateau at 23–26 mya (Knesel et al., 2008) at a period when Australia was warm and humid (Martin, 2006; Byrne et al., 2008). Therefore, the invasion of a tropically adapted, ancestral Gehyra from the Melanesian region at this time is plausible and supported by our ancestral state reconstruction analysis. In contrast with the other Australian Gekkotan lineages which have a Gondwanan origin, the divergence between Australian and Melanesian Gehyra is more recent (Gamble et al., 2008b; Oliver and Sanders, 2009) as is consequently the diversification within Australian Gehyra.
Hypothesis 2—tropically adapted and arid-adapted species complexes
All of our analyses find two clades within the Australian radiation, consistent with previous molecular studies (Sistrom et al., 2009, Oliver et al., 2010). The content of our two groups mostly matches the subdivision proposed by Mitchell (1965) and King; however, two of King’s australis group species, G. occidentalis and G. xenopus, fall into our variegata clade. Species contained within the initial concepts of the G. australis clade (Figure 2) were, on average, larger-bodied taxa (King, 1983; Horner, 2005) associated with the tropical, subtropical and monsoonal tropics of Australia and southern New Guinea, whereas the variegata clade comprised smaller bodied species associated with the arid and semi-arid zones (King, 1979; Moritz, 1986). Both G. occidentalis and G. xenopus are relatively large bodied (maximum SVL (Snout-vent length) >65 mm), both are confined to the monsoonal Kimberley region of Western Australia and both branch near the base of the G. variegata clade. Although it is true that many of the members of the G. variegata clade are smaller bodied than those in the G. australis clade, body size appears to be somewhat labile in this group, with larger species branching close to smaller species (Sistrom et al., 2012). The one consistent aspect of body size appears to be that the smallest species (maximum SVL<45 mm) are confined to the variegata group, but no general conclusion applies to medium and larger body sizes. Similarly, the tropical–arid dichotomy is weakened by the likely plesiomorphic nature of tropical adaptations and the fact that the G. variegata clade includes tropical species.
Hypothesis 3—evaluation of chromosomal speciation patterns
King (1984)hypothesized that the diversification of the Australian Gehyra was driven by chromosomal speciation and proposed a detailed evolutionary scenario by which this may have occurred. However, this scenario came under considerable scrutiny (Sites and Moritz, 1987) owing to the inconclusive nature of assumptions regarding the allopatric distribution of chromosome races and reproductive isolation between them. A prediction of King’s (1984) proposed evolutionary scenario is that reproductively isolated chromosome races should change in a predictable fashion as one moves from the root of the tree towards the tips, with changes at speciation points. It is clear from the distribution of chromosome races in our analysis (Figure 2) and the results of ancestral state reconstruction analysis that this is not the case. Furthermore, the placement of G. occidentalis in the G. variegata clade means that no G. australis clade members are now known to have a 2n=44 karyotype. Therefore, the assumption that the 2n=44 chromosome karyotype is the ancestral state of the Australian Gehyra is questionable and not supported by our ancestral state reconstruction. Given our phylogeny, either the independent evolution of karyotypes (such as 2n=42a) or reversal (to 2n=44) are necessary to explain the observed karyotypes, but neither phenomenon was countenanced in King’s model. King’s work undoubtedly revealed the fact of large-scale cryptic speciation in Gehyra, but the mechanism he proposed has not proven to be a sufficient explanation.
Data archiving
Data available from the Dryad Digital Repository: doi:10.5061/dryad.7t354 and from GenBank: accession numbers KJ025084–KJ025522.
References
Bertozzi T, Sanders KL, Sistrom MJ, Gardner MG . (2012). Anonymous nuclear loci in non-model organisms: making the most of high-throughput genome surveys. Bioinformatics 28: 1807–1810.
Burbrink FT, Pyron AR . (2011). The impact of gene-tree/species tree discordance on diversification rate estimation. Evolution 65: 1851–1861.
Byrne M, Yeates DK, Joseph L, Kearney M, Bowler J, Williams MAJ et al. (2008). Birth of a biome: insights into the assembly and maintenance of Australian arid zone biota. Mol Ecol 20: 4398–4417.
Chung Y, Ané C . (2011). Comparing two Bayesian methods for gene tree/species tree reconstruction: Simulations with incomplete lineage sorting and horizontal gene transfer. Syst Biol 60: 261–275.
Cranston KA . (2010). Summarizing gene tree incongruence at multiple phylogenetic depths. In Knowles LL, Kubatko LS (eds). Estimating Species Trees: Practical and Theoretical Aspects. Wiley-Blackwell: New York, NY, USA. pp 129–142.
Creevey CJ, McInerney JO . (2002). An algorithm for detecting directional and non-directional positive selection, neutrality and negative selection in protein coding DNA sequences. Gene 300: 43–51.
Creevey CJ, McInerney JO . (2003). CRANN: detecting adaptive evolution in protein-coding DNA sequences. Bioinformatics 19: 1726.
Degnan JH, Rosenberg NA . (2009). Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol 24: 332–340.
Drummond AJ, Rambaut A . (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214.
Drummond AJ, Ho SYW, Phillips MJ, Rambaut A . (2006). Relaxed phylogenetics and dating with confidence. PLoS Biol 4: e88.
Drummond AJ, Rambaut A . (2010) BEAST v1.6. Available from http://beast.bio.ed.ac.uk/ Last accessed 26 April 2011.
Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C et al. (2010) Geneious v5.3. Available from www.geneious.com.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acid Res 32: 1792–1797.
Edwards SV, Liu L, Pearl DK . (2007). High-resolution species trees without concatenation. Proc Natl Acad Sci USA 104: 5936–5941.
Edwards SV . (2009). Is a new and general theory of molecular systematics emerging? Evolution 63: 1–19.
Felsenstein J . (2005) PHYLIP (Phylogeny Inference Package) version 3: 6. Distributed by the author. Department of Genome Sciences, University of Washington: Seattle, WA, USA.
Fujita MK, McGuire JA, Donnellan SC, Moritz CM . (2010). Diversification and persistence at the arid-monsoonal interface: Australia-wide biogeography of the Bynoe’s gecko (Heteronotia binoei: Gekkonidae). Evolution 64: 2293–2314.
Gamble TP, Bauer AM, Greenbaum E, Jackman TR . (2008a). Out of the blue: a novel trans-Atlantic clade of geckos (Gekkota, Squamata). Zool Scirpta 37: 355–366.
Gamble TP, Bauer AM, Greenbaum E, Jackman TR . (2008b). Evidence for Gondwanan vicariance in an ancient clade of gecko lizards. J Biogeogr 35: 88–104.
Gamble TP, Bauer AM, Colli GR, Greenbaum E, Jackman TR, Vitt LJ et al. (2010). Coming to America: multiple origins of New World geckos. J Evol Biol 24: 231–244.
Han D, Zhou K, Bauer AM . (2004). Phylogenetic relationships among gekkotan lizards inferred from C-mos nuclear DNA sequences and a new classification of the Gekkota. Biol J Linn Soc 83: 353–368.
Heled J, Drummond AJ . (2010). Bayesian inference of species trees from multilocus data. Mol Biol Evol 27: 570–580.
Horner PP . (2005). Gehyra koira sp. nov. (Reptilia: Gekkonidae), a new species of lizard with two allopatric subspecies from the Ord-Victoria region of north-western Australia and a key to the Gehyra australis species complex. Beagle 21: 165–174.
Huang H, He Q, Kubatko LS, Knowles LL . (2010). Sources of error inherent in species-tree estimation: Impact of mutational and coalescent effects on accuracy and implications for choosing among different methods. Syst Biol 59: 573–583.
King M . (1979). Karyotypic evolution in Gehyra (Gekkonidae: Reptilia) I. The Gehyra variegata-punctata complex. Aust J Zool 27: 373–393.
King M . (1982). Karyotypic evolution in Gehyra (Gekkonidae: Reptilia) II. A new species from the Alligator Rivers region in northern Australia. Aust J Zool 30: 93–101.
King M . (1983). Karyotypic evolution in Gehyra (Gekkonidae: Reptilia) III. The Gehyra australis species complex. Aust J Zool 31: 723–741.
King M . (1984). Karyotypic evolution in Gehyra (Gekkonidae: Reptilia) IV. Chromosome change and speciation. Genetica 64: 101–114.
Knesel KM, Cohen BE, Vasconcelos PM, Thiede DS . (2008). Rapid change in drift of the Australian plate records collision with Ontong Java plateau. Nature 454: 754–758.
Knowles LL, Carstens BC . (2007). Delimiting species without monophyletic gene trees. Syst Biol 56: 887–895.
Kruskal WH, Wallis WA . (1952). Use of ranks in one-criterion variance analysis. J Am Stat Assoc 47: 583–621.
Kubatko LS, Degnan JH . (2007). Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst Biol 56: 17–24.
Kubatko LS, Carstens BC, Knowles LL . (2009). STEM: Species tree estimation using maximum likelihood for gene trees under coalescence. Bioinformatics 25: 971–973.
Lanier HC, Knowles LL . (2012). Is recombination a problem for species tree analysis? Syst Biol 61: 691–701.
Larget BR, Kotha SK, Dewey CN, Ané C . (2010). BUCKy: gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics 26: 2910–2911.
Leaché AD . (2009). Species tree discordance traces to phylogeographic clade boundaries in North American fence lizards (Sceloporus). Syst Biol 58: 547–559.
Liu L, Pearl DK . (2007). Species trees from gene trees: reconstructing Bayesian posterior distributions of a species phylogeny using estimated gene tree distributions. Syst Biol 56: 504–514.
Liu L, Yu L . (2011). Estimating species trees from unrooted gene trees. Syst Biol 60: 661–667.
Maddison WP . (1997). Gene trees in species trees. Syst Biol 46: 523–536.
Maddison WP, Knowles LL . (2006). Inferring phylogeny despite incomplete lineage sorting. Syst Biol 55: 21–30.
Martin HA . (2006). Cenozoic climatic changes and the development of the arid vegetation of Australia. J Arid Environ 66: 533–563.
McCormack JE, Heled J, Delaney KS, Peterson AT, Knowles LL . (2010). Calibrating divergence times on species trees versus gene trees: implications for speciation history of Aphelocoma jays. Evolution 65: 184–202.
Mitchell FJ . (1965). Australian geckos assigned to the genus Gehyra Gray (Reptilia: Gekkonidae). Senckenb Biol 46: 287–319.
Molnar RE . (2000). The long and honorable history of monitors and their kin. In: Pianka ER, King DR, King RA (eds). Varanoid Lizards of the World. Indiana University Press: Bloomington and Indianapolis,. pp 10–67.
Moritz CM . (1984). The evolution of a highly variable sex chromosome in Gehyra purpurascens (Gekkonidae). Chromosoma 90: 111–119.
Moritz CM . (1986). The population biology of Gehyra (Gekkonidae): chromosome change and speciation. Syst Biol 35: 46–67.
Moritz CM . (1992). The population biology of Gehyra (Gekkonidae) III. Patterns of microgeographic variation. J Evol Biol 5: 661–676.
Oliver PM, Sanders KL . (2009). Molecular evidence for Gondwanan origins of multiple lineages within a diverse Australasian gecko radiation. J Biogeogr 36: 2044–2055.
Oliver PM, Sistrom MJ, Tjaturadi B, Krey K, Richards SJ . (2010). On the status and relationships of the gecko species Gehyra barea (Kopstein 1926), with description of new specimens and a range extension. Zootaxa 2354: 45–55.
Pepper M, Doughty D, Hutchinson M, Keogh S . (2011). Ancient drainages divide cryptic species in Australia’s arid zone: morphological and multi-gene evidence for four new species of beaked geckos (Rhyncoedura). Mol Phylogenet Evol 61: 810–822.
Pinho C, Rocha S, Carvalho BM, Lopes S, Mourao S, Vallinoto M et al. (2010). New primes for the amplification and sequencing of nuclear loci in a taxonomically wide set of reptiled and amptibians. Conservation Genet. Resource 2: 181–185.
Posada D . (2008). jModelTest: phylogenetic model averaging. Mol Biol Evol 25: 1253–1256.
R Development Core Team. (2008) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org.
Rambaut A . (2009) Figtree v1.4. Available from http://tree.bio.ed.ac.uk/ . Last accessed 28 May 2011..
Reid N, Demboski JR, Sullivan J . (2012). Phylogeny estimation of the radiation of western North American chipmunks (Tamias) in the face of introgression using reproductive protein genes. Syst Biol 61: 44–62.
Revell LJ . (2012). Phytools: an R package for phylogenetic comparative biology (and other things). Method Ecol Evol 3: 217–223.
Ronquist F, Huelsenbeck JP . (2003). MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Rowe KC, Aplin KP, Baverstock PR, Moritz CM . (2010). Recent and rapid speciation with limited morphological diversity in the genus Rattus. Syst Biol 60: 188–203.
Rozas J . (2009). DNA sequence polymorphism analysis using DnaSP. In: Posada D (ed). Bioinformatics for DNA Sequence Analysis; Methods. In Molecular Biology Series. Humana Press: New Jersey, USA vol. 537, pp 337–350.
Russell AP, Bauer AM . (2002). Underwood’s classification of the geckos: A 21st century appreciation. Bull Br Mus (Nat Hist) Zool 68: 113–121.
Salichos L, Rokas A . (2013). Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497: 327–331.
Sanders KL, Lee MSY . (2007). Evaluating molecular clock calibrations using Bayesian analyses with soft and hard bounds. Biol Lett 3: 275–279.
Sanders KL, Lee MSY, Leys R, Foster R, Keogh SJ . (2008). Molecular phylogeny and divergence dates for Australasian elapids and sea snakes (Hydrophiinae): evidence from seven genes for rapid evolutionary radiations. J Evol Biol 21: 682–695.
Sistrom M, Donnellan SC, Hutchinson MN . (2013). Delimiting species in recent radiations with low levels of morphological diver: a case study in Australian Gehyra geckos. Mol Phylogenet Evol 68: 135–143.
Sistrom MJ, Hutchinson MN, Hutchinson RG, Donnellan SC . (2009). Molecular phylogeny of Australian Gehyra (Squamata: Gekkonidae) and taxonomic revision of Gehyra variegata in south-eastern Australia. Zootaxa 2277: 14–32.
Sistrom MJ, Edwards DL, Donnellan SC, Hutchinson MN . (2012). Morphological differentiation correlates with ecological but not genetic divergence in a Gehyra gecko. J Evol Biol 25: 647–660.
Sites JR, Moritz CM . (1987). Chromosomal evolution and speciation revisited. Syst Biol 36: 153–174.
Stephens M, Smith N, Donnelly P . (2001). A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68: 978–989.
Townsend JP . (2007). Profiling phylogenetic informativeness. Syst Biol 56: 222–231.
Wiens JJ, Morrill MC . (2011). Missing data in phylogenetic analysis: reconciling results from simulations and empirical data. Syst Biol 60: 719–731.
Wiens JJ, Brandley MC, Reeder TW . (2006). Why does a trait evolve multiple times within a clade? Repeated evolution of snake like boby form in squamate reptiles. Evol 60: 114–123.
Acknowledgements
This work was funded by an ABRS Grant 207-43 awarded to MN.H and SCD. We thank Kate Sanders and Mike Lee for advice on suitable calibrations for divergence estimation, Scott Edwards for advice on sampling design and Hailey Lanier and Danielle Edwards for reviewing and improving the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Rights and permissions
About this article
Cite this article
Sistrom, M., Hutchinson, M., Bertozzi, T. et al. Evaluating evolutionary history in the face of high gene tree discordance in Australian Gehyra (Reptilia: Gekkonidae). Heredity 113, 52–63 (2014). https://doi.org/10.1038/hdy.2014.6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2014.6