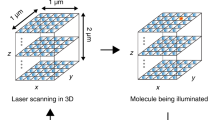
Abstract
Monitoring the behavior of single molecules in living cells is a powerful approach to investigate the details of cellular processes. Owing to their optical, chemical and biofunctional properties, semiconductor quantum dot (QD) probes promise to be tools of choice in this endeavor. Here we review recent advances that allow ever more controlled experiments at the single-nanoparticle level in live cells. Several examples, related to membrane dynamics, cell signaling or intracellular transport, illustrate how single QD tracking can be readily used to decipher complex biological processes and address key concepts that underlie cellular organization and dynamics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Saxton, M.J. & Jacobson, K. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26, 373–399 (1997).
Joo, C., Balci, H., Ishitsuka, Y., Buranachai, C. & Ha, T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 77, 51–76 (2008).
Bruchez, M., Moronne, M., Gin, P., Weiss, S. & Alivisatos, A.P. Semiconductor nanocrystals as fluorescent biological labels. Science 281, 2013–2016 (1998).
Dahan, M. et al. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science 302, 442–445 (2003). First paper describing QD use to track single receptors in live cells.
Walter, N.G., Huang, C.Y., Manzo, A.J. & Sobhy, M.A. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat. Methods 5, 475–489 (2008).
Selvin, P.R. & Ha, T. Single-molecule techniques: a laboratory manual (Cold Spring Harbor Laboratory Press, 2008).
Wieser, S. & Schutz, G.J. Tracking single molecules in the live cell plasma membrane-do's and don't's. Methods 46, 131–140 (2008).
Weiss, S. Fluorescence spectroscopy of single biomolecules. Science 283, 1676–1683 (1999).
Lord, S.J., Lee, H.L. & Moerner, W.E. Single-molecule spectroscopy and imaging of biomolecules in living cells. Anal. Chem. advance online publication, doi:10.1021/ac9024889 (17 February 2010).
Haggie, P.M., Kim, J.K., Lukacs, G.L. & Verkman, A.S. Tracking of quantum dot-labeled CFTR shows near immobilization by C-terminal PDZ interactions. Mol. Biol. Cell 17, 4937–4945 (2006).
Resch-Genger, U., Grabolle, M., Cavaliere-Jaricot, S., Nitschke, R. & Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 5, 763–775 (2008).
Pons, T. & Mattoussi, H. Investigating biological processes at the single molecule level using luminescent quantum dots. Ann. Biomed. Eng. 37, 1934–1959 (2009).
Stefani, F.D., Hoogenboom, J.P. & Barkai, E. Beyond quantum jumps: blinking nanoscale light emitters. Phys. Today 62, 34–39 (2009).
Alivisatos, A.P. Semiconductor clusters, nanocrystals and quantum dots. Science 271, 933–937 (1996).
Reiss, P., Protiere, M. & Li, L. Core/shell semiconductor nanocrystals. Small 5, 154–168 (2009).
Michalet, X. et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science 307, 538–544 (2005). A review of QD properties and applications for biological imaging.
Bentzen, E.L. et al. Surface modification to reduce nonspecific binding of quantum dots in live cell assays. Bioconjug. Chem. 16, 1488–1494 (2005).
Liu, W. et al. Compact biocompatible quantum dots functionalized for Cellular imaging. J. Am. Chem. Soc. 130, 1274–1284 (2008).
Doose, S., Tsay, J.M., Pinaud, F. & Weiss, S. Comparison of photophysical and colloidal properties of biocompatible semiconductor nanocrystals using fluorescence correlation spectroscopy. Anal. Chem. 77, 2235–2242 (2005).
Howarth, M. et al. Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nat. Methods 5, 397–399 (2008). With reference 40, describes clever engineering of small and monofunctional QDs, a substantial improvement for targeting and tracking single membrane proteins.
Groc, L. et al. Surface trafficking of neurotransmitter receptor: comparison between single-molecule/quantum dot strategies. J. Neurosci. 27, 12433–12437 (2007).
Nechyporuk-Zloy, V., Dieterich, P., Oberleithner, H., Stock, C. & Schwab, A. Dynamics of single potassium channel proteins in the plasma membrane of migrating cells. Am. J. Physiol. Cell Physiol. 294, C1096–C1102 (2008).
Dubertret, B. et al. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science 298, 1759–1762 (2002).
Roullier, V. et al. High-affinity labeling and tracking of individual histidine-tagged proteins in live cells using Ni2+ Tris-nitrilotriacetic acid quantum dot conjugates. Nano Lett. 9, 1228–1234 (2009).
Lees, E.E., Nguyen, T.L., Clayton, A.H.A., Mulvaney, P. & Muir, B.W. The preparation of colloidally stable, water-soluble, biocompatible, semiconductor nanocrystals with a small hydrodynamic diameter. Acs Nano 3, 1121–1128 (2009).
Murcia, M.J., Minner, D.E., Mustata, G.M., Ritchie, K. & Naumann, C.A. Design of quantum dot-conjugated lipids for long-term, high-speed tracking experiments on cell surfaces. J. Am. Chem. Soc. 130, 15054–15062 (2008).
Aldana, J., Wang, Y.A. & Peng, X.G. Photochemical instability of CdSe nanocrystals coated by hydrophilic thiols. J. Am. Chem. Soc. 123, 8844–8850 (2001).
Susumu, K. et al. Enhancing the stability and biological functionalities of quantum dots via compact multifunctional ligands. J. Am. Chem. Soc. 129, 13987–13996 (2007).
Pinaud, F., King, D., Moore, H.P. & Weiss, S. Bioactivation and cell targeting of semiconductor CdSe/ZnS nanocrystals with phytochelatin-related peptides. J. Am. Chem. Soc. 126, 6115–6123 (2004).
Dif, A. et al. Small and stable peptidic PEGylated quantum dots to target polyhistidine-tagged proteins with controlled stoichiometry. J. Am. Chem. Soc. 131, 14738–14746 (2009).
Pinaud, F. et al. Dynamic partitioning of a glycosyl-phosphatidylinositol-anchored protein in glycosphingolipid-rich microdomains imaged by single-quantum dot tracking. Traffic 10, 691–712 (2009).
Iyer, G. et al. High affinity scFv-hapten pair as a tool for quantum dot labeling and tracking of single proteins in live cells. Nano Lett. 8, 4618–4623 (2008).
Bannai, H., Levi, S., Schweizer, C., Dahan, M. & Triller, A. Imaging the lateral diffusion of membrane molecules with quantum dots. Nat. Protocols 1, 2628–2634 (2006).
Crane, J.M. & Verkman, A.S. Long-range nonanomalous diffusion of quantum dot-labeled aquaporin-1 water channels in the cell plasma membrane. Biophys. J. 94, 702–713 (2008).
Ehrensperger, M.V., Hanus, C., Vannier, C., Triller, A. & Dahan, M. Multiple association states between glycine receptors and gephyrin identified by SPT analysis. Biophys. J. 92, 3706–3718 (2007).
Lidke, D.S., Lidke, K.A., Rieger, B., Jovin, T.M. & Arndt-Jovin, D.J. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J. Cell Biol. 170, 619–626 (2005).
Gralle, M., Botelho, M.G. & Wouters, F.S. Neuroprotective secreted amyloid precursor protein acts by disrupting amyloid precursor protein dimers. J. Biol. Chem. 284, 15016–15025 (2009).
Andrews, N.L. et al. Actin restricts Fc epsilon RI diffusion and facilitates antigen-induced receptor immobilization. Nat. Cell Biol. 10, 955–963 (2008).
Marks, K.M. & Nolan, G.P. Chemical labeling strategies for cell biology. Nat. Methods 3, 591–596 (2006).
Howarth, M., Takao, K., Hayashi, Y. & Ting, A.Y. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc. Natl. Acad. Sci. USA 102, 7583–7588 (2005).
Genin, E. et al. CrAsH—quantum dot nanohybrids for smart targeting of proteins. J. Am. Chem. Soc. 130, 8596–8597 (2008).
So, M.-K., Yao, H. & Rao, J. HaloTag protein-mediated specific labeling of living cells with quantum dots. Biochem. Biophys. Res. Commun. 374, 419–423 (2008).
Kim, J. et al. Ni-nitrilotriacetic acid-modified quantum dots as a site-specific labeling agent of histidine-tagged proteins in live cells. Chem. Commun. 2008, 1910–1912 (2008).
Swift, J.L. & Cramb, D.T. Nanoparticles as fluorescence labels: is size all that matters? Biophys. J. 95, 865–876 (2008).
Bonasio, R. et al. Specific and covalent labeling of a membrane protein with organic fluorochromes and quantum dots. Proc. Natl. Acad. Sci. USA 104, 14753–14758 (2007).
George, N., Pick, H., Vogel, H., Johnsson, N. & Johnsson, K. Specific labeling of cell surface proteins with chemically diverse compounds. J. Am. Chem. Soc. 126, 8896–8897 (2004).
Sunbul, M., Yen, M., Zou, Y. & Yin, J. Enzyme catalyzed site-specific protein labeling and cell imaging with quantum dots. Chem. Commun. 2008, 5927–5929 (2008).
Zanchet, D., Micheel, C.M., Parak, W.J., Gerion, D. & Alivisatos, A.P. Electrophoretic isolation of discrete Au nanocrystal/DNA conjugates. Nano Lett. 1, 32–35 (2001).
Sperling, R.A., Pellegrino, T., Li, J.K., Chang, W.H. & Parak, W.J. Electrophoretic separation of nanoparticles with a discrete number of functional groups. Adv. Funct. Mater. 16, 943–948 (2006).
Meijering, E., Smal, I. & Danuser, G. Tracking in molecular bioimaging. IEEE Signal Process. Mag. 23, 46–53 (2006).
Thompson, R.E., Larson, D.R. & Webb, W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 82, 2775–2783 (2002).
Cheezum, M.K., Walker, W.F. & Guilford, W.H. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophys. J. 81, 2378–2388 (2001).
Bonneau, S., Dahan, M. & Cohen, L.D. Single quantum dot tracking based on perceptual grouping using minimal paths in a spatiotemporal volume. IEEE Trans. Image Process. 14, 1384–1395 (2005).
Serge, A., Bertaux, N., Rigneault, H. & Marguet, D. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods 5, 687–694 (2008).
Jaqaman, K. et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702 (2008). With reference 54, this paper describes freely downloadable software for tracking single nanoparticles in image sequences.
Qian, H., Sheetz, M.P. & Elson, E.L. Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys. J. 60, 910–921 (1991).
Bouzigues, C. & Dahan, M. Transient directed motions of GABA(A) receptors in growth cones detected by a speed correlation index. Biophys. J. 92, 654–660 (2007).
Huet, S. et al. Analysis of transient behavior in complex trajectories: application to secretory vesicle dynamics. Biophys. J. 91, 3542–3559 (2006).
Meilhac, N., Le Guyader, L., Salome, L. & Destainville, N. Detection of confinement and jumps in single-molecule membrane trajectories. Phys. Rev. E 73, 011915 (2006).
Simson, R., Sheets, E.D. & Jacobson, K. Detection of temporary lateral confinement of membrane proteins using single-particle tracking analysis. Biophys. J. 69, 989–993 (1995).
Helmuth, J.A., Burckhardt, C.J., Koumoutsakos, P., Greber, U.F. & Sbalzarini, I.F. A novel supervised trajectory segmentation algorithm identifies distinct types of human adenovirus motion in host cells. J. Struct. Biol. 159, 347–358 (2007).
Masson, J.B. et al. Inferring maps of forces inside cell membrane microdomains. Phys. Rev. Lett. 102, 048103 (2009).
Durisic, N. et al. Detection and correction of blinking bias in image correlation transport measurements of quantum dot tagged macromolecules. Biophys. J. 93, 1338–1346 (2007).
Lidke, D.S. et al. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat. Biotechnol. 22, 198–203 (2004). Earliest use of ligand-conjugated QDs for the real-time imaging of EGF-erb/HER signal transduction in live cells.
Bouzigues, C., Morel, M., Triller, A. & Dahan, M. Asymmetric redistribution of GABA receptors during GABA gradient sensing by nerve growth cones analyzed by single quantum dot imaging. Proc. Natl. Acad. Sci. USA 104, 11251–11256 (2007).
O'Connell, K.M.S., Rolig, A.S., Whitesell, J.D. & Tamkun, M.M. Kv2.1 potassium channels are retained within dynamic cell surface microdomains that are defined by a perimeter fence. J. Neurosci. 26, 9609–9618 (2006).
Tamkun, M.M., O'Connell, K.M.S. & Rolig, A.S. A cytoskeletal-based perimeter fence selectively corrals a sub-population of cell surface Kv2.1 channels. J. Cell Sci. 120, 2413–2423 (2007).
Bates, I.R. et al. Membrane lateral diffusion and capture of CFTR within transient confinement zones. Biophys. J. 91, 1046–1058 (2006).
Chen, H., Titushkin, I., Stroscio, M. & Cho, M. Altered membrane dynamics of quantum dot-conjugated integrins during osteogenic differentiation of human bone marrow derived progenitor cells. Biophys. J. 92, 1399–1408 (2007).
Kodippili, G.C. et al. Imaging of the diffusion of single band 3 molecules on normal and mutant erythrocytes. Blood 113, 6237–6245 (2009).
Crane, J.M., Van Hoek, A.N., Skach, W.R. & Verkman, A.S. Aquaporin-4 dynamics in orthogonal arrays in live cells visualized by quantum dot single particle tracking. Mol. Biol. Cell 19, 3369–3378 (2008).
Crane, J.M. & Verkman, A.S. Determinants of aquaporin-4 assembly in orthogonal arrays revealed by live-cell single-molecule fluorescence imaging. J. Cell Sci. 122, 813–821 (2009).
Heine, M. et al. Surface mobility of postsynaptic AMPARs tunes synaptic transmission. Science 320, 201–205 (2008).
Groc, L. et al. NMDA receptor surface mobility depends on NR2A–2B subunits. Proc. Natl. Acad. Sci. USA 103, 18769–18774 (2006).
Levi, S. et al. Homeostatic regulation of synaptic GlyR numbers driven by lateral diffusion. Neuron 59, 261–273 (2008).
Mikasova, L., Groc, L., Choquet, D. & Manzoni, O.J. Altered surface trafficking of presynaptic cannabinoid type 1 receptor in and out synaptic terminals parallels receptor desensitization. Proc. Natl. Acad. Sci. USA 105, 18596–18601 (2008).
Geng, L., Qian, Y.K., Madhavan, R. & Peng, H.B. Transmembrane mechanisms in the assembly of the postsynaptic apparatus at the neuromuscular junction. Chem. Biol. Interact. 175, 108–112 (2008).
Triller, A. & Choquet, D. New concepts in synaptic biology derived from single-molecule imaging. Neuron 59, 359–374 (2008).
Charrier, C., Ehrensperger, M.-V., Dahan, M., Levi, S. & Triller, A. Cytoskeleton regulation of glycine receptor number at synapses and diffusion in the plasma membrane. J. Neurosci. 26, 8502–8511 (2006).
Groc, L. et al. NMDA receptor surface trafficking and synaptic subunit composition are developmentally regulated by the extracellular matrix protein reelin. J. Neurosci. 27, 10165–10175 (2007).
Renner, M., Choquet, D. & Triller, A. Control of the postsynaptic membrane viscosity. J. Neurosci. 29, 2926–2937 (2009).
Zhang, Q., Cao, Y.Q. & Tsien, R.W. Quantum dots provide an optical signal specific to full collapse fusion of synaptic vesicles. Proc. Natl. Acad. Sci. USA 104, 17843–17848 (2007).
Zhang, Q., Li, Y.L. & Tsien, R.W. The dynamic control of kiss-and-run and vesicular reuse probed with single nanoparticles. Science 323, 1448–1453 (2009). With reference 82, an original use of single QD optical properties and size to address the controversial issues of kiss and run versus full collapse vesicular fusion as a mechanism for presynaptic release in neurons.
Cui, B. et al. One at a time, live tracking of NGF axonal transport using quantum dots. Proc. Natl. Acad. Sci. USA 104, 13666–13671 (2007).
Echarte, M.M., Bruno, L., Arndt-Jovin, D.J., Jovin, T.M. & Pietrasanta, L.I. Quantitative single particle tracking of NGF-receptor complexes: transport is bidirectional but biased by longer retrograde run lengths. FEBS Lett. 581, 2905–2913 (2007).
Rajan, S.S., Liu, H.Y. & Vu, T.Q. Ligand-bound quantum dot probes for studying the molecular scale dynamics of receptor endocytic trafficking in live cells. ACS Nano. 2, 1153–1166 (2008).
Joo, K.I. et al. Site-specific labeling of enveloped viruses with quantum dots for single virus tracking. ACS Nano. 2, 1553–1562 (2008).
Courty, S., Luccardini, C., Bellaiche, Y., Cappello, G. & Dahan, M. Tracking individual kinesin motors in living cells using single quantum-dot imaging. Nano Lett. 6, 1491–1495 (2006). First report of QD use for intracellular tracking, with a study of individual kinesin motor movements in the cytoplasm of live cells.
Pierobon, P. et al. Velocity, processivity, and individual steps of single myosin V molecules in live cells. Biophys. J. 96, 4268–4275 (2009).
Nelson, S.R., Ali, M.Y., Trybus, K.M. & Warshaw, D.M. Random walk of processive, quantum dot-labeled myosin Va molecules within the actin cortex of COS-7 cells. Biophys. J. 97, 509–518 (2009).
Keren, K., Yam, P.T., Kinkhabwala, A., Mogilner, A. & Theriot, J.A. Intracellular fluid flow in rapidly moving cells. Nat. Cell Biol. 11, 1219–U137 (2009).
Ishihama, Y. & Funatsu, T. Single molecule tracking of quantum dot-labeled mRNAs in a cell nucleus. Biochem. Biophys. Res. Commun. 381, 33–38 (2009).
Wells, N.P. et al. Going beyond 2D: following membrane diffusion and topography in the IgE-Fc[epsilon]RI system using 3-dimensional tracking microscopy. Proc. SPIE 7185, 71850Z-1–71850Z-13 (2009).
Holtzer, L., Meckel, T. & Schmidt, T. Nanometric three-dimensional tracking of individual quantum dots in cells. Appl. Phys. Lett. 90, 053902-1–053902-3 (2007).
Ram, S., Prabhat, P., Chao, J., Ward, E.S. & Ober, R.J. High accuracy 3D quantum dot tracking with multifocal plane microscopy for the study of fast intracellular dynamics in live cells. Biophys. J. 95, 6025–6043 (2008).
Thompson, M.A., Lew, M.D., Badieirostami, M. & Moerner, W.E. Localizing and tracking single nanoscale emitters in three dimensions with high spatiotemporal resolution using a double-helix point spread function. Nano Lett. 10, 211–218 (2010).
Choi, H.S. et al. Renal clearance of quantum dots. Nat. Biotechnol. 25, 1165–1170 (2007).
Smith, A.M. & Nie, S. Minimizing the hydrodynamic size of quantum dots with multifunctional multidentate polymer ligands. J. Am. Chem. Soc. 130, 11278–11279 (2008).
Xie, R., Battaglia, D. & Peng, X. Colloidal InP nanocrystals as efficient emitters covering blue to near-infrared. J. Am. Chem. Soc. 129, 15432–15433 (2007).
Zimmer, J.P. et al. Size series of small indium arsenide-zinc selenide core-shell nanocrystals and their application to in vivo imaging. J. Am. Chem. Soc. 128, 2526–2527 (2006).
Hohng, S. & Ha, T. Near-complete suppression of quantum dot blinking in ambient conditions. J. Am. Chem. Soc. 126, 1324–1325 (2004).
Chen, Y. et al. Giant multishell CdSe nanocrystal quantum dots with suppressed blinking. J. Am. Chem. Soc. 130, 5026–5027 (2008).
Mahler, B. et al. Towards non-blinking colloidal quantum dots. Nat. Mater. 7, 659–664 (2008).
Wang, X. et al. Non-blinking semiconductor nanocrystals. Nature 459, 686–689 (2009).
Misteli, T. The concept of self-organization in cellular architecture. J. Cell Biol. 155, 181–185 (2001).
Huang, B., Bates, M. & Zhuang, X. Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 78, 993–1016 (2009).
Ulbrich, M.H. & Isacoff, E.Y. Subunit counting in membrane-bound proteins. Nat. Methods 4, 319–321 (2007).
Delehanty, J., Mattoussi, H. & Medintz, I. Delivering quantum dots into cells: strategies, progress and remaining issues. Anal. Bioanal. Chem. 393, 1091–1105 (2009).
Jaiswal, J.K., Mattoussi, H., Mauro, J.M. & Simon, S.M. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat. Biotechnol. 21, 47–51 (2003).
Derfus, A.M., Chan, W.C.W. & Bhatia, S.N. Intracellular delivery of quantum dots for live cell labeling and organelle tracking. Adv. Mater. 16, 961–966 (2004).
Delehanty, J.B. et al. Self-assembled quantum dot-peptide bioconjugates for selective intracellular delivery. Bioconjug. Chem. 17, 920–927 (2006).
Ruan, G., Agrawal, A., Marcus, A.I. & Nie, S. Imaging and tracking of Tat peptide-conjugated quantum dots in living cells: new insights into nanoparticle uptake, intracellular transport, and vesicle shedding. J. Am. Chem. Soc. 129, 14759–14766 (2007).
Chan, W.C.W. & Nie, S.M. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 281, 2016–2018 (1998).
Tekle, C., v Deurs, B., Sandvig, K. & Iversen, T.-G. Cellular trafficking of quantum dot-ligand bioconjugates and their induction of changes in normal routing of unconjugated ligands. Nano Lett. 8, 1858–1865 (2008).
Yoo, J., Kambara, T., Gonda, K. & Higuchi, H. Intracellular imaging of targeted proteins labeled with quantum dots. Exp. Cell Res. 314, 3563–3569 (2008).
Kim, B.Y.S. et al. Biodegradable quantum dot nanocomposites enable live cell labeling and imaging of cytoplasmic targets. Nano Lett. 8, 3887–3892 (2008).
Duan, H. & Nie, S. Cell-penetrating quantum dots based on multivalent and endosome-disrupting surface coatings. J. Am. Chem. Soc. 129, 3333–3338 (2007).
Qi, L. & Gao, X. Quantum dot-amphipol nanocomplex for intracellular delivery and real-time imaging of siRNA. ACS Nano 2, 1403–1410 (2008).
Jablonski, A.E., Humphries, W.H. & Payne, C.K. Pyrenebutyrate-mediated delivery of quantum dots across the plasma membrane of living cells. J. Phys. Chem. B 113, 405–408 (2009).
Chen, X., Kis, A., Zettl, A. & Bertozzi, C.R. A cell nanoinjector based on carbon nanotubes. Proc. Natl. Acad. Sci. USA 104, 8218–8222 (2007).
Park, S., Kim, Y.-S., Kim, W.B. & Jon, S. Carbon nanosyringe array as a platform for intracellular delivery. Nano Lett. 9, 1325–1329 (2009).
Acknowledgements
We thank Y. Bellaiche, G. Cappello, P. Desbiolles, V. Marchi-Artzner, M. Renner and A. Triller for careful reading of the manuscript and valuable discussions. F.P. acknowledges financial support from a Marie Curie-Intra-European Fellowship (contract MEIF-CT-2006-040210) and a European Molecular Biology Organization Long-Term Fellowship. S.C. is supported by postdoctoral fellowships from Université Pierre et Marie Curie and the Fondation pour la Recherche Médicale. A.S. is supported by a doctoral fellowship from the Marie Curie Training Network. M.D. acknowledges funding from the Fondation pour la Recherche Médicale, Agence Nationale pour la Recherche (ANR-05-PNANO-045), the Human Frontier Science Program (RGP0005/2007) and the Centre C'Nano Ile de France.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Pinaud, F., Clarke, S., Sittner, A. et al. Probing cellular events, one quantum dot at a time. Nat Methods 7, 275–285 (2010). https://doi.org/10.1038/nmeth.1444
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1444
This article is cited by
-
Single-virus tracking with quantum dots in live cells
Nature Protocols (2023)
-
Tracking single particles for hours via continuous DNA-mediated fluorophore exchange
Nature Communications (2021)
-
Improved resolution in single-molecule localization microscopy using QD-PAINT
Experimental & Molecular Medicine (2021)
-
Methods for Intracellular Delivery of Quantum Dots
Topics in Current Chemistry (2021)
-
Organic Nanocrystals Based on a Solid-emission-tunable AIEgen for Cell Imaging
Chemical Research in Chinese Universities (2021)