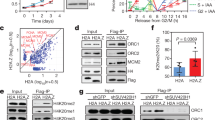
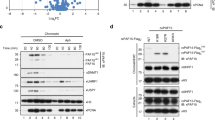
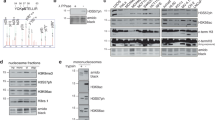
Abstract
The recognition of distinctly modified histones by specialized ‘effector’ proteins constitutes a key mechanism for transducing molecular events at chromatin to biological outcomes1. Effector proteins influence DNA-templated processes, including transcription, DNA recombination and DNA repair; however, no effector functions have yet been identified within the mammalian machinery that regulate DNA replication. Here we show that ORC1—a component of ORC (origin of replication complex), which mediates pre-DNA replication licensing2—contains a bromo adjacent homology (BAH) domain that specifically recognizes histone H4 dimethylated at lysine 20 (H4K20me2). Recognition of H4K20me2 is a property common to BAH domains present within diverse metazoan ORC1 proteins. Structural studies reveal that the specificity of the BAH domain for H4K20me2 is mediated by a dynamic aromatic dimethyl-lysine-binding cage and multiple intermolecular contacts involving the bound peptide. H4K20me2 is enriched at replication origins, and abrogating ORC1 recognition of H4K20me2 in cells impairs ORC1 occupancy at replication origins, ORC chromatin loading and cell-cycle progression. Mutation of the ORC1 BAH domain has been implicated in the aetiology of Meier–Gorlin syndrome (MGS)3,4, a form of primordial dwarfism5, and ORC1 depletion in zebrafish results in an MGS-like phenotype4. We find that wild-type human ORC1, but not ORC1–H4K20me2-binding mutants, rescues the growth retardation of orc1 morphants. Moreover, zebrafish depleted of H4K20me2 have diminished body size, mirroring the phenotype of orc1 morphants. Together, our results identify the BAH domain as a novel methyl-lysine-binding module, thereby establishing the first direct link between histone methylation and the metazoan DNA replication machinery, and defining a pivotal aetiological role for the canonical H4K20me2 mark, via ORC1, in primordial dwarfism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Taverna, S. D., Li, H., Ruthenburg, A. J., Allis, C. D. & Patel, D. J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature Struct. Mol. Biol. 14, 1025–1040 (2007)
Duncker, B. P., Chesnokov, I. N. & McConkey, B. J. The origin recognition complex protein family. Genome Biol. 10, 214 (2009)
Bicknell, L. S. et al. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nature Genet. 43, 356–359 (2011)
Bicknell, L. S. et al. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nature Genet. 43, 350–355 (2011)
Klingseisen, A. & Jackson, A. P. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 25, 2011–2024 (2011)
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007)
Callebaut, I., Courvalin, J. C. & Mornon, J. P. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 446, 189–193 (1999)
Onishi, M., Liou, G. G., Buchberger, J. R., Walz, T. & Moazed, D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell 28, 1015–1028 (2007)
Sampath, V. et al. Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes. Mol. Cell. Biol. 29, 2532–2545 (2009)
Armache, K. J., Garlick, J. D., Canzio, D., Narlikar, G. J. & Kingston, R. E. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 334, 977–982 (2011)
Bua, D. J. et al. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS ONE 4, e6789 (2009)
Bell, S. P., Mitchell, J., Leber, J., Kobayashi, R. & Stillman, B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83, 563–568 (1995)
Dorn, E. S. & Cook, J. G. Nucleosomes in the neighborhood: new roles for chromatin modifications in replication origin control. Epigenetics 6, 552–559 (2011)
Bell, S. P. & Dutta, A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374 (2002)
Zhang, Z., Hayashi, M. K., Merkel, O., Stillman, B. & Xu, R. M. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 21, 4600–4611 (2002)
Botuyan, M. V. et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 (2006)
Li, H. et al. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol. Cell 28, 677–691 (2007)
Schotta, G. et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 22, 2048–2061 (2008)
Noguchi, K., Vassilev, A., Ghosh, S., Yates, J. L. & DePamphilis, M. L. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J. 25, 5372–5382 (2006)
Tardat, M. et al. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nature Cell Biol. 12, 1086–1093 (2010)
Tardat, M., Murr, R., Herceg, Z., Sardet, C. & Julien, E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J. Cell Biol. 179, 1413–1426 (2007)
Miotto, B. & Struhl, K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 22, 2633–2638 (2008)
Kitsberg, D., Selig, S., Keshet, I. & Cedar, H. Replication structure of the human β-globin gene domain. Nature 366, 588–590 (1993)
Guernsey, D. L. et al. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nature Genet. 43, 360–364 (2011)
Sun, X. J. et al. Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS ONE 3, e1499 (2008)
Costas, C. et al. Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks. Nature Struct. Mol. Biol. 18, 395–400 (2011)
Miotto, B. & Struhl, K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol. Cell 37, 57–66 (2010)
Jacob, Y. et al. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature 466, 987–991 (2010)
Brustel, J., Tardat, M., Kirsh, O., Grimaud, C. & Julien, E. Coupling mitosis to DNA replication: the emerging role of the histone H4-lysine 20 methyltransferase PR-Set7. Trends Cell Biol. 21, 452–460 (2011)
Oda, H. et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol. Cell. Biol. 29, 2278–2295 (2009)
Shi, X. et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–99 (2006)
Matthews, A. G. et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450, 1106–1110 (2007)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Mendez, J. & Stillman, B. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 (2000)
Kuo, A. J. et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell 44, 609–620 (2011)
Acknowledgements
We thank R. Tennen for critical reading of the manuscript. This work was supported in part by grants to O.G. (R01 GM079641), D.J.P. (Abby Rockefeller Mauze, STARR and Maloris Foundations) and J.K.C. (DP1 OD003792), and a predoctoral fellowship to A.J.K. (Genentech Foundation). O.G. is a recipient of an Ellison Senior Scholar in Aging Award.
Author information
Authors and Affiliations
Contributions
A.J.K. and P.C. performed the molecular biology, cellular and zebrafish studies; J.S. performed structural and binding affinity studies; S.Y. and J.K.C. advised on zebrafish experiments; S.I.-M. assisted in protein production and crystallization. A.J.K., P.C., J.S., D.J.P. and O.G. designed studies, analysed data and wrote the paper. All authors discussed and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-11 and Supplementary Table 1. (PDF 1855 kb)
Rights and permissions
About this article
Cite this article
Kuo, A., Song, J., Cheung, P. et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier–Gorlin syndrome. Nature 484, 115–119 (2012). https://doi.org/10.1038/nature10956
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10956
This article is cited by
-
Decoding chromatin states by proteomic profiling of nucleosome readers
Nature (2024)
-
ORC1 binds to cis-transcribed RNAs for efficient activation of replication origins
Nature Communications (2023)
-
TNRC18 engages H3K9me3 to mediate silencing of endogenous retrotransposons
Nature (2023)
-
Distinct roles of Arabidopsis ORC1 proteins in DNA replication and heterochromatic H3K27me1 deposition
Nature Communications (2023)
-
The expanding genetic and clinical landscape associated with Meier-Gorlin syndrome
European Journal of Human Genetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.