Abstract
Autosomal recessive mental retardation (AR-MR) may account for up to 25% of genetic mental retardation (MR). So far, mapping of AR-MR genes in consanguineous families has resulted in six nonsyndromic genes, whereas more than 2000 genes might contribute to AR-MR. We propose to use outbred families with multiple affected siblings for AR-MR gene identification. Homozygosity mapping in ten outbred families with affected brother–sister pairs using a 250 K single nucleotide polymorphism array revealed on average 57 homozygous regions over 1 Mb in size per affected individual (range 20–74). Of these, 21 homozygous regions were shared between siblings on average (range 8–36). None of the shared regions of homozygosity (SROHs) overlapped with the nonsyndromic genes. A total of 13 SROHs had an overlap with previously reported loci for AR-MR, namely with MRT8, MRT9, MRT10 and MRT11. Among these was the longest observed SROH of 11.0 Mb in family ARMR1 on chromosome 19q13, which had 2.9 Mb (98 genes) in common with the 5.4 Mb MRT11 locus (195 genes). These data support that homozygosity mapping in outbred families may contribute to identification of novel AR-MR genes.
Similar content being viewed by others
Introduction
Mental retardation (MR), also referred to as intellectual disability, is a common neurodevelopmental disorder affecting approximately 1–3% of the general population.1 Clinically, DSM-IV defines MR as significant subaverage intellectual functioning – intelligence quotients below 70 – with an onset before the age of 18 years and impairment in adaptive functioning such as self care, social and interpersonal skills and work (American Psychiatric Association (1994), DMS-IV). MR can be subdivided into syndromic and nonsyndromic forms based on the presence or absence of additional features, although this distinction is clinically not always obvious. The etiology of MR is heterogeneous and despite recent improvement of cytogenetic and molecular technologies, less than 50% of patients have an etiological diagnosis in clinical practice, hampering medical care and prognosis of the patient and genetic counseling of the families.2, 3
Genetic causes contribute significantly to MR and among these autosomal recessive inheritances may account for a substantial part of this disorder. Although no recent estimations have been made, autosomal recessive genes have previously been estimated to account for up to 25% of unexplained MR.4, 5, 6, 7 This is more than two times as frequent as the contribution of single X-chromosomal genes to MR, which is estimated to explain MR in 10% of affected men.8, 9, 10 For practical reasons, over the last few decades, the focus in genetic MR research has been on X-linked MR, leading to the identification of 90 disease genes on the X-chromosome of which 38 (42%) lead to nonsyndromic MR.11 In contrast, of the 348 genes contributing to autosomal recessive mental retardation (AR-MR) phenotypes, only six genes (1.6%) have been identified giving rise to nonsyndromic AR-MR (OMIM: http://www.ncbi.nlm.nih.gov/omim). On the basis of the number of MR genes on the X chromosome − 11% of X-chromosome protein-coding genes are implicated in X-linked MR – we estimate that there are about 2000 AR-MR genes (11% of all 18 625 autosomal protein-coding genes) ((Vega v37; September 2009); http://vega.sanger.ac.uk/Homo_sapiens/Info/Index). The limited number of nonsyndromic AR-MR genes has mostly been elucidated through studies of consanguineous families, large enough to perform linkage analysis, resulting in significant LOD scores. As these families are rare and most often originate from geographical regions where consanguineous marriages are common, other, parallel approaches for the identification of genes causing this heterogeneous condition are required. Recent technological advances, such as whole genome homozygosity mapping with high-resolution single nucleotide polymorphism (SNP) arrays combined with next generation sequencing, enable the analysis of outbred families (brother–sister or sister–sister pairs), which are more common than the consanguineous families used thus far. Homozygosity mapping in such small, outbred families is performed under the assumption that a homozygous mutation in a recessive disease gene is passed on to the affected child by both parents who received the mutant allele from a common ancestor. Previous studies in other and less heterogeneous autosomal recessive disorders, such as retinitis pigmentosa, steroid resistant nephrotic syndrome and nephronopthisis have shown the applicability of homozygosity mapping for identification of mutations in families and isolated patients without consanguinity.12, 13, 14 The percentage of homozygous mutations in these cohorts might be as high as 70%.14 Here, we report the first study of homozygosity mapping in outbred MR families.
Patients and methods
Families
A total of 10 families with brother–sister pairs with MR were included in this study (in total 22 individuals). Eight families consisted of two affected individuals. In two families, three individuals were analyzed: in family ARMR1 a brother–sister pair with an unaffected brother and in family ARMR10 monozygotic twin brothers and their sister. Patients were clinically evaluated by a clinical geneticist in the human genetics department of the Radboud University Nijmegen Medical Centre in Nijmegen, the Netherlands (Supplementary Table 1). The study was approved by the medical ethics committee of the Radboud University Nijmegen Medical Centre.
Homozygosity mapping
Patient DNA was isolated from lymphocytes as described by Miller et al.15 Samples were hybridized on an Affymetrix NspI 250 K SNP array. SNP array experiments were performed according to the manufacturer's protocols (Affymetrix, Santa Clara, CA, USA). Copy number estimates were determined using the CNAG software package (v2.0)16 to exclude causative copy number aberrations. Genotypes were called by Affymetrix Genotype Console Software v2.1. All hybridizations had >85% successful genotype calls.
PLINK v1.06 was used to systematically identify runs of homozygous called SNPs in the 22 siblings, 19 MR patients from consanguineous parents, 817 MR patients from non-consanguineous parents and 159 healthy controls from an outbred population.17 In each window of 50 SNPs, up to five SNPs with a missing call and a maximum of two heterozygous called SNPs were allowed. We determined (1) regions of at least 1 Mb that contained a minimum of 50 contiguous, genotyped homozygous SNPs, (2) regions ≥1.5 Mb and containing a minimum 75 contiguous SNPs, (3) regions ≥2 Mb and containing a minimum of 100 SNPs and (4) regions ≥5 Mb and containing a minimum of 250 SNPs. For allelic matching, segments were compared pairwise, and samples were grouped into the same haplotype group if at least 95% of jointly nonmissing, SNPs were identical. Regions that were homozygous in both sibs, but having different haplotypes, were excluded.
Results and discussion
Clinical data
We studied 10 families with mild-to-severe MR (families ARMR1-10, Supplementary Table 1). All families had one affected man and one affected woman, except for ARMR10. This family had an affected woman and two affected men, the latter two being monozygotic twins. Additional clinical features were observed in eight families, ranging from minor clinical features, such as pectus carinatum in ARMR4, to major clinical features, such as hypokinetic rigid syndrome with severe autism in ARMR10.
Homozygosity mapping
Individual regions of homozygosity
Population studies often use a size cutoff value for the detection of regions of homozygosity (ROHs) of 1 Mb.18, 19, 20, 21 Homozygous mutations in outbred families have been identified in regions as small as 2.1 Mb and mutations in consanguineous families are usually found in large linkage intervals encompassing several to tens of megabases.14, 22 Therefore, we choose to analyze the 10 outbred families with four different ROH size cutoff values, namely 1, 1.5, 2 and 5 Mb. The number of ROHs over 1 Mb in size varied from 20 in the male patient of ARMR3 to 74 in the male patient of ARMR1, and was on average 57 per individual (Table 1 ). We observed on average 14 ROHs over 1.5 Mb (range 2–22) and 4 ROHs over 2 Mb (range 0–9) in size. ROHs over 5 Mb in size were observed in three families (ARMR1, 7 and 8). Notably, the female patient of ARMR7 has seven ROHs considerably larger than 5 Mb (range 6.4–27.1 Mb), indicating that in this family the parents are likely related, although her affected brother has only one homozygous stretch of 6 Mb. The average of 57 ROHs per individual is in line with previous reports showing on average 31 ROHs in healthy individuals (range 0–115 in 2429 individuals).18, 19, 20
To test whether the siblings were truly from outbred families, we compared the total amount of genomic homozygosity (all regions >1 Mb) of the siblings with (1) 19 MR patients from consanguineous parents, (2) 817 MR patients from non-consanguineous parents and (3) 159 healthy controls from an outbred population. With Student's t-test, the siblings having on average 178 Mb (SD: 75 Mb) of the genome homozygous, differed significantly (P=8.34 E-08) from the consanguineous MR patients, who had on average 358 Mb (SD: 98 Mb) of their genome homozygous. There was no significant difference between the siblings and the non-consanguineous MR patients (P=0.441, average homozygosity 191 Mb, SD: 49 Mb) or the healthy controls (P=0.337, average homozygosity 194 Mb, SD: 74 Mb). This shows that the total amount of genomic homozygosity of the siblings, in the current study, is not different from that observed in an outbred population.
Shared regions of homozygosity
We observed shared regions of homozygosity (SROHs) in the siblings in all 10 families and categorized these SROHs according to sizes longer than 1, 1.5, 2 and 5 Mb, respectively (Figure 1). On average, siblings shared 21 regions longer than 1 Mb (range: 8–36), which represents almost 40% of the individual ROHs per sib (Table 1). Siblings shared on average four regions over 1.5 Mb in size (average: 27% of individual ROHs) and one region over 2 Mb in size (average: 25% of individual ROHs). Two families, ARMR1 and 8, showed one single SROH longer than 5 Mb; 11.0 Mb on chromosome 19q13 and 8.4 Mb on chromosome 6q23, respectively. On the basis of Mendelian inheritance, siblings are expected to share 25% of their individual ROHs. Therefore, the 40% overlap of ROHs longer than 1 Mb observed between sibs is most likely partly due to false-positive ROHs. However, the rate of false-positive ROHs drops as the size of the ROHs increases, as we observed for the 1.5 and 2 Mb regions with 27 and 25% homozygous regions shared between siblings, respectively.
This graph shows the number of shared regions of homozygosity between siblings (SROHs) in each family, categorized by different size cutoff values for ROH detection (ROH size >1, >1.5, >2, >5 Mb). The number of SROHs of 1 Mb or longer varies from eight SROHs in ARMR3 to 36 in ARMR6 (mean 21), this number is reduced to on average four SROHs of 1.5 Mb or longer, whereas only two families, ARMR1 and ARMR8 have an SROH over 5 Mb in size. Black: SROHs with size cutoff for ROH analysis of >1 Mb, dark grey: SROHs size cutoff >1.5 Mb, light gray: SROHs size cutoff >2 Mb, white: SROHs size cutoff >5 Mb.
In ARMR1, consisting of four siblings (an affected brother and sister and two healthy brothers), we also genotyped one of the healthy brothers. In this family, 18 ROHs longer than 1 Mb were shared between the patients but not with the healthy sib, thereby reducing the number of candidate regions by 55%. Notably, the longest SROH of 11.0 Mb on chromosome 19q13 was heterozygous in the healthy brother.
For all SROHs over 2 Mb in size (12 in total), we compared haplotypes of the ARMR families with haplotypes of the 817 non-consanguineous MR patients and the 159 healthy controls to observe whether these haplotypes were unique or part of a frequently occurring haplotype. We considered a haplotype as shared between individuals when there was a minimal overlap of 2 Mb. Three SROHs had a unique haplotype (ARMR1, chr16: 78.198.864–80.847.224; ARMR8, chr6: 130.485.817–138.854.223 and chr9: 131.487.840–133.745.038, http://genome.ucsc.edu/, hg18), among which the second longest SROH of 8.4 Mb on chromosome 6 in ARMR8. The 11.0 Mb SROH of family ARMR1 (chr19: 38.737.515–48.316.888) had a 2.3 Mb overlap with one patient (chr19: 387.37.515–41.057.851), the haplotype of the remaining 8.7 Mb of this 11.0 Mb SROH showed no overlap. Another SROH of 2.6 Mb on chromosome 6 in ARMR9 had an overlap with three patients (chr6: 62.030.184–64.647.424). Six SROHs had a more common haplotype occurring in 2.6–14.4% of the 976 studied individuals (Supplementary Table 3).
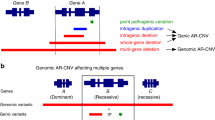
Shared regions of homozygosity and known AR-MR loci
None of the SROHs encompassed one of the six reported nonsyndromic AR-MR genes or encompassed syndromic AR-MR genes that could explain the phenotype in these families. Overlapping of SROHs with known AR-MR loci was observed for MRT7 in ARMR1 and 2, for MRT8 in ARMR2, 3, 6 and 7, for MRT9 in ARMR7, for MRT10 in ARMR1, 6, 7, 8 and 10, and for MRT11 in ARMR1 (OMIM: %611093, %611094, %611095, %611096, 611097) (Figure 2, Supplementary Table 2). The size of these homozygous segments varied from 950 kb to 1.9 Mb, except for the 11.0 Mb segment that overlaps with MRT11 (Figure 2). Families ARMR2, 3 and 6 shared the same haplotype in the MRT8 locus (1.1 Mb overlap: rs16929951; rs16930750, 15 genes) as did ARMR1, 6, 7, 8 and 10 for the MRT10 locus. As more than 2000 genes might contribute to AR-MR and the mutation frequency of each single gene is presumably below 0.1%, each family in this study will most probably have a unique genetic defect, giving rise to the MR. Therefore, the overlap of several families with part of the MRT7, MRT8 and MRT10 loci, is unlikely to contain the causative genetic defect in all families, but likely to reflect common ROHs as reported by Lencz et al21. In their study, 9 and 15% of 144 healthy individuals were homozygous for MRT8 and MRT10, respectively. Of more interest is the overlap of the single families with the MRT9 and MRT11 loci. Especially the latter, as this overlap of 2.9 Mb with MRT11 is part of an 11.0 Mb homozygous region (388 SNPs: rs1864132; rs16959168) that is shared between the affected siblings of family 1 and is heterozygous in the unaffected brother. This 11.0 Mb ROH is the only homozygous stretch exceeding 5 Mb in both siblings. The MRT11 locus (MIM: %611097), reported by Najmabadi et al,22 was mapped in a consanguineous Iranian family with four patients with nonsyndromic, moderate MR, with a maximum LOD score of 4.0. The MRT11 candidate region is a 5.4 Mb region between rs2109075 and rs8101149 containing 195 genes, whereas the 2.9 Mb overlap reduces the number of candidate genes to 98 (Map Viewer, build 36.3, www.ncbi.nlm.nih.gov/mapview). The results obtained for ARMR1, with nonsyndromic, severe MR, support that homozygosity mapping in outbred families might contribute to identification of novel AR-MR genes especially in combination with next generation sequencing technologies.
SROHs showing overlap with MRT7–10 (http://genome.ucsc.edu/, hg18). (a) Overlap of the MRT7 locus with ARMR1 and ARMR2. (b) Overlap of the MRT8 locus with ARMR2, 3, 6 and 7. The first three families share the same haplotype. This region is also reported by Lencz et al21 to be homozygous in 9% of 144 healthy individuals. (c) Overlap of the MRT9 locus with ARMR7. (d) Overlap of the MRT10 locus with ARMR1, 6, 7, 8, 10. All families share the same haplotype. This region is reported to be homozygous in 15% of 144 healthy individuals by Lencz et al.21 (e) 11 Mb SROH of ARMR1, showing 2.9 Mb overlap with the MRT11 locus. (f) Enlargement of the 2.9 Mb overlap containing 98 genes.
We report the first study of homozygosity mapping in outbred MR families to map AR-MR genes. Our data of 10 AR-MR families, show that most outbred families share only relatively short homozygous regions (<5 Mb) with only two individual families sharing one relatively long homozygous region (8.4 and 11.0 Mb). Follow-up studies in these and other families combining data of SROHs with next generation sequencing will further show whether homozygosity mapping in outbred families is a useful approach to unravel the molecular basis of AR-MR.
References
Leonard H, Wen X : The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev 2002; 8: 117–134.
Stevenson RE, Procopio-Allen AM, Schroer RJ, Collins JS : Genetic syndromes among individuals with mental retardation. Am J Med Genet A 2003; 123A: 29–32.
Rauch A, Hoyer J, Guth S et al: Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A 2006; 140A: 2063–2074.
Bartley JA, Hall BD : Mental retardation and multiple congenital anomalies of unknown etiology: frequency of occurrence in similarly affected sibs of the proband. Birth Defects Orig Artic Ser 1978; 14: 127–137.
Chelly J, Khelfaoui M, Francis F, Cherif B, Bienvenu T : Genetics and pathophysiology of mental retardation. Eur J Hum Genet 2006; 14: 701–713.
Priest JH, Thuline HC, Laveck GD, Jarvis DB : An approach to genetic-factors in mental retardation – studies of families containing at least 2 siblings admitted to a state institution for the retarded. Am J Ment Defic 1961; 66: 42–50.
Wright SW, Tarjan G, Eyer L : Investigation of families with 2 or more mentally defective siblings – clinical observations. Am J Dis Child 1959; 97: 445–463.
de Brouwer AP, Yntema HG, Kleefstra T et al: Mutation frequencies of X-linked mental retardation genes in families from the EuroMRX consortium. Hum Mutat 2007; 28: 207–208.
Mandel JL, Chelly J : Monogenic X-linked mental retardation: is it as frequent as currently estimated? The paradox of the ARX (Aristaless X) mutations. Eur J Hum Genet 2004; 12: 689–693.
Ropers HH, Hamel BCJ : X-linked mental retardation. Nat Rev Genet 2005; 6: 46–57.
Gecz J, Shoubridge C, Corbett M : The genetic landscape of intellectual disability arising from chromosome X. Trends Genet 2009; 25: 308–316.
Abd El-Aziz MM, Barragan I, O’Driscoll CA et al: EYS, encoding an ortholog of Drosophila spacemaker, is mutated in autosomal recessive retinitis pigmentosa. Nat Genet 2008; 40: 1285–1287.
Collin RW, Littink KW, Klevering BJ et al: Identification of a 2 Mb human ortholog of Drosophila eyes shut/spacemaker that is mutated in patients with retinitis pigmentosa. Am J Hum Genet 2008; 83: 594–603.
Hildebrandt F, Heeringa SF, Ruschendorf F et al: A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet 2009; 5: e1000353.
Miller SA, Dykes DD, Polesky HF : A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Nannya Y, Sanada M, Nakazaki K et al: A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res 2005; 65: 6071–6079.
Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Curtis D, Vine AE, Knight J : Study of regions of extended homozygosity provides a powerful method to explore haplotype structure of human populations. Ann Hum Genet 2008; 72: 261–278.
Gibson J, Morton NE, Collins A : Extended tracts of homozygosity in outbred human populations. Hum Mol Genet 2006; 15: 789–795.
Nalls MA, Simon-Sanchez J, Gibbs JR et al: Measures of autozygosity in decline: globalization, urbanization, and its implications for medical genetics. Plos Genetics 2009; 5: e1000415.
Lencz T, Lambert C, DeRosse P et al: Runs of homozygosity reveal highly penetrant recessice loci in schizophrenia. Proc Natl Acad Sci USA 2007; 104: 19942–19947.
Najmabadi H, Motazacker MM, Garshasbi M et al: Homozygosity mapping in consanguineous families reveals extreme heterogeneity of non-syndromic autosomal recessive mental retardation and identifies 8 novel gene loci. Hum Genet 2007; 121: 43–48.
Acknowledgements
We are grateful to the patients and their families for their support and cooperation. This work has been supported by grants from the Dutch Organisation for Health Research and Development (ZON-MW) (917-86-319 to BBAdV), Hersenstichting Nederland (BBAdV).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Schuurs-Hoeijmakers, J., Hehir-Kwa, J., Pfundt, R. et al. Homozygosity mapping in outbred families with mental retardation. Eur J Hum Genet 19, 597–601 (2011). https://doi.org/10.1038/ejhg.2010.167
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2010.167
Keywords
This article is cited by
-
Genetics of intellectual disability in consanguineous families
Molecular Psychiatry (2019)
-
Genome-wide estimates of inbreeding in unrelated individuals and their association with cognitive ability
European Journal of Human Genetics (2014)