Abstract
The target of rapamycin complex 1 (TORC1) integrates various hormonal and nutrient signals to regulate cell growth, proliferation, and differentiation. Amino acid-dependent activation of TORC1 is mediated via the yeast EGO complex (EGOC) consisting of Gtr1, Gtr2, Ego1, and Ego3. Here, we identify the previously uncharacterized Ycr075w-a/Ego2 protein as an additional EGOC component that is required for the integrity and localization of the heterodimeric Gtr1-Gtr2 GTPases, equivalent to mammalian Rag GTPases. We also report the crystal structure of the Ego1-Ego2-Ego3 ternary complex (EGO-TC) at 2.4 Å resolution, in which Ego2 and Ego3 form a heterodimer flanked along one side by Ego1. Structural data also reveal the structural conservation of protein components between the yeast EGO-TC and the human Ragulator, which acts as a GEF for Rag GTPases. Interestingly, however, artificial tethering of Gtr1-Gtr2 to the vacuolar membrane is sufficient to activate TORC1 in response to amino acids even in the absence of the EGO-TC. Our structural and functional data therefore support a model in which the EGO-TC acts as a scaffold for Rag GTPases in TORC1 signaling.
Similar content being viewed by others
Introduction
The target of rapamycin complex 1 (TORC1) is a highly conserved protein kinase complex that couples a vast range of external stimuli to cell growth and metabolism1,2. Activated TORC1 positively regulates anabolic processes (such as ribosome biogenesis and translation initiation) and inhibits catabolic processes (such as macroautophagy) via the phosphorylation of well-characterized downstream effectors, including the S6 kinase 1 and the type 2A and 2A-related phosphatases. Among the wide range of signals that impinge on TORC1 activity, amino acids are particularly potent activators3,4,5. The mechanism by which this signal is transduced to TORC1 is largely conserved and requires the Rag family GTPases in both yeast and mammals3,6.
In mammalian cells, RagA or RagB forms a heterodimeric complex with RagC or RagD and is active when RagA/B is in its GTP-bound state and RagC/D is bound with GDP7,8. The active Rag heterodimer binds the TORC1 subunit Raptor to promote its recruitment to the lysosomal membranes where it is in a position to interact with its activator GTPase Rheb7. The Rag GTPase heterodimers are themselves recruited to lysosomes via the Ragulator complex: a multisubunit complex that exerts guanine nucleotide-exchange factor (GEF) activity towards RagA/B7,9. The Ragulator complex consists of the lysosome membrane anchor LAMTOR1/p18, a heterodimer comprised of LAMTOR2/p14 and LAMTOR3/MP1, and two newly identified subunits LAMTOR4/C7orf59 and LAMTOR5/HBXIP9. Together, these components influence the nucleotide loading state of RagA/B, promoting the active state of the heterodimer10. Structural studies of the Ragulator components have shown that LAMTOR2 and LAMTOR3 form a heterodimer whereas LAMTOR5 alone can exist as a homodimer11,12,13. However, how LAMTOR1-5 are assembled into the Ragulator complex is unclear.
The equivalent components of the Rag GTPases and Ragulator in yeast, collectively referred to as the EGO complex (EGOC), are also vital for amino acid-dependent activation of TORC114. The EGOC comprises the Rag family GTPases Gtr1 and Gtr2 (yeast equivalents of RagA/B and RagC/D, respectively), in addition to the Ego1/Meh1 and Ego3/Slm4 proteins that serve to anchor the complex to the vacuolar membrane15. Amino acids (particularly leucine) promote the GTP-bound state of Gtr1, which, in combination with the GDP-bound Gtr2, interacts with and activates TORC114,16. Like LAMTOR1/p18, Ego1 is anchored to the membrane of the vacuole through an N-terminal lipid modification whereas Ego3 can form a homodimer that is structurally similar to the heterodimer of LAMTOR2-LAMTOR3 and is required for the recruitment of the Gtr GTPases to the vacuole17,18,19,20.
In yeast, it is unclear whether the known EGOC components also have the capacity to promote the activated state of the Gtr1-Gtr2 heterodimer or whether the recruitment of additional components to the EGOC may fulfill this function. Indeed, a number of Gtr regulators have previously been described, including the Gtr1 GEF Vam614, the positive Gtr1 regulator leucyl-tRNA synthetase21, and the SEACIT and SEACAT complexes that also regulate the activity of Gtr115,22. Moreover, no obvious counterparts of mammalian LAMTOR4 and LAMTOR5, shown to be essential for the GEF activity of the Ragulator, appeared to exist in yeast. However, a recent study that employed HHpred, a highly sensitive method for structural homology detection, identified two uncharacterized yeast proteins Ycr075w-a and Ynr034w-a as hypothetical LAMTOR4 and LAMTOR5 homologs23. While very little is known about either protein, the NMR structure of Ynr034w-a reveals the presence of a Roadblock domain-like fold, a common feature of many GTPase-interacting proteins. Curiously, Roadblock domains are predicted to be present not only in Ycr075w-a and the LAMTOR4-LAMTOR5 heterodimer, but also to exist in the LAMTOR2-LAMTOR3 heterodimer, the Ego3 homodimer, as well as within the C-terminal dimerization domains of the Rag/Gtr GTPases23.
Here, we investigate the functions of Ycr075w-a and Ynr034w-a (hereafter referred to as Ego2 and Ego4, respectively) and identify Ego2 as an additional component of the TORC1 signaling pathway in S. cerevisiae. We demonstrate that Ego2 interacts directly with Ego1 and Ego3 to form a ternary complex required for the proper localization of the Gtr GTPases at the vacuolar membrane and subsequent TORC1 activation. We also report the crystal structure of the Ego1-Ego2-Ego3 complex, which provides new insights into the architecture of the EGOC and the molecular basis for its functional role in promoting Rag GTPase-dependent TORC1 signaling.
Results
Ego2 is a positive regulator of TORC1 signaling
The observations made by Levine et al.23 indicated that the uncharacterized yeast proteins Ycr075w-a/Ego2 and/or Ynr034w-a/Ego4 might, like LAMTOR4-LAMTOR5, be positive regulators of the Gtr GTPases and TORC1. To investigate the potential roles of these two proteins in TORC1 signaling, we assessed the ability of cells lacking either Ego2 or Ego4 to grow in the presence of increasing concentrations of the TORC1 inhibitor rapamycin. Like for cells lacking either of the Gtr GTPases, ego2Δ cells were hypersensitive to sub-inhibitory concentrations of rapamycin (3 ng/ml and 5 ng/ml) and failed to recover after exposure to rapamycin (Figure 1A). In contrast, cells lacking Ego4 grew comparably to wild-type (WT) cells, and cells deleted for both EGO2 and EGO4 exhibited similar rapamycin sensitivity to that of single ego2Δ deletion cells. The combination of deleting EGO2 together with either GTR1 or GTR2 did not result in additive rapamycin sensitivity, suggesting that Ego2 may act within the same pathway as the EGOC to promote TORC1 activity. Interestingly, cells deleted for both GTR2 and EGO4 exhibited a stronger growth defect on rapamycin when compared to the single-deletion mutants (Figure 1A). To verify these observations, we assessed TORC1 activity by monitoring the phosphorylation of the TORC1 target Sch924 (Figure 1B). Loss of Ego2 resulted in consistently lower TORC1 activity when compared to WT cells, whereas no reduction in the level of Sch9 phosphorylation could be detected in cells lacking Ego4. In contrast with the effect of its loss, overproduction of Ego2 did not significantly affect TORC1 activity (Figure 1C). Similarly, overproduction of Ego4 resulted in no significant alteration in TORC1 activity.
Ego2 promotes TORC1 activity. (A) Cells deleted for the indicated genes were serially (10-fold) diluted and spotted onto solid YPD medium containing 0, 3, or 5 ng/ml rapamycin and incubated at 30 °C for 3 days. For rapamycin recovery, cells were treated with 200 ng/ml rapamycin for 6 h before spotting onto solid YPD medium. (B) Immunoblots detecting the extent of phosphorylation within the C-terminus of Sch9 were used to quantify TORC1 activity (the ratio of hyperphosphorylated (+P)/hypophosphorylated (−P) Sch9) in the indicated deletion strains. The values (means ± SD; n = 3) were normalized to WT cells and presented in the bar graph. One representative immunoblot is shown. It is worth noting that overall Sch9 protein levels are consistently reduced in strains exhibiting low TORC1 activity, suggesting that long-term TORC1 inactivation influences the regulation of Sch9. (C) Sch9 phosphorylation was monitored in cells in which Ego2 or Ego4 was overexpressed under the control of the ADH1 promoter. The indicated TORC1 activity values were calculated as in B. (D, E) Bar graph representing TORC1 activity as measured in B in the presence of the indicated Gtr1 or Gtr2 alleles (D). Representative immunoblots showing the extent of Sch9 phosphorylation are shown in E. (F) Serial 10-fold dilutions of cells expressing the different Gtr1 and Gtr2 alleles from their endogenous promoters were spotted onto synthetic defined medium containing 0 or 5 ng/ml rapamycin and grown at 30 °C for 3 days.
We next tested whether Ego2 affected TORC1 activity through the EGOC. To this end, we combined ego2Δ with overexpression of signaling-competent or signaling-compromised Gtr1 and Gtr2 alleles. Neither the absence of Gtr1 or Gtr2, nor overexpression of signaling-compromised Gtr1S20L (which has a low affinity for nucleotides) or Gtr2Q66L(GTP-locked), further reduced the low TORC1 activity of ego2Δ cells (Figure 1D and 1E), consistent with the notion that Ego2 and the Gtr GTPases function in a common pathway. However, the elevated TORC1 activity levels observed in cells overexpressing active Gtr1Q65L or Gtr2S23L were strongly dependent on Ego2 (Figure 1D and 1E). Moreover, expression of Gtr1Q65L or Gtr2S23L from their endogenous promoters could not alleviate the rapamycin hypersensitivity observed in ego2Δ cells (Figure 1F). Since expression of neither Gtr1Q65L nor Gtr2S23L could rescue TORC1 activity in the absence of Ego2, and overexpression of EGO2 did not lead to significantly higher levels of TORC1 activity, Ego2 is unlikely to function solely to promote the signaling-proficient nucleotide-binding states of the Gtr GTPases. Instead, Ego2 may affect the function of the EGOC via another mechanism or act in parallel to the EGOC to promote TORC1 activity. The loss of Ego4 did not impair the effects of the different Gtr1 or Gtr2 variants on TORC1 activity and rapamycin sensitivity (Figure 1D-1F), except in the presence of the active form of Gtr2S23L where hyperactivation of TORC1 was blunted in ego4Δ cells (Figure 1D and 1E). The genetic interaction between EGO4 and GTR2 is the only phenotype linked to EGO4 so far and its biological significance remains unclear.
Ego2 is essential for the assembly of the EGOC at the vacuolar membrane
The localization of the EGOC at the vacuolar periphery is essential for its function in TORC1 activation. We previously demonstrated that the vacuolar localization of the Gtr1-Gtr2 heterodimer is dependent on Ego3, while Ego1 is necessary to anchor the entire complex to the membrane20. To investigate whether the phenotypes observed in ego2Δ cells resulted from a perturbation of the EGOC itself, we analyzed the subcellular localization of Ego1 and Ego3. The localization of Ego1-GFP was unchanged in strains lacking Ego2, Ego3, Ego4, or both Ego2 and Ego3 together (Figure 2A), consistent with its known role as a vacuolar anchor for the complex. In contrast, Ego3-GFP failed to localize to the vacuole in the absence of Ego2, but not Ego4 (Figure 2A). This is similar to the localization pattern observed in the absence of Ego1 and demonstrates that, like Ego1, Ego2 is necessary for the recruitment of Ego3 at the vacuolar membrane. We then asked whether Ego2 was therefore necessary for the interaction between Ego1 and Ego3. In the presence of Ego2, Ego1-TAP co-precipitated well with Ego3-GFP (Figure 2B). In contrast, the interaction between Ego1-TAP and Ego3-GFP was entirely lost in cells lacking Ego2, while the absence of Ego4 had no effect. Ego2 is therefore critical for the interaction between Ego1 and Ego3. Given the importance of Ego2 for Ego3 recruitment, we assumed that it would also be essential for the proper localization of Gtr1 and Gtr2. Indeed, the vacuolar localization of Gtr1-GFP and Gtr2-GFP was severely reduced in the absence of Ego2 and this could not be rescued by expression of their signaling-competent forms (Figure 2C). To further examine the effect of loss of Ego2 on the Gtr1-Gtr2 heterodimer, we analyzed the interaction between Gtr1 and Gtr2. Interestingly, a reduced amount of Gtr2 co-precipitated with Gtr1 in ego2Δ cells, and this was also the case in cells lacking either Ego1 or Ego3 (Figure 2D). To exclude that this destabilization of the heterodimer in ego2Δ cells resulted purely from changes in the nucleotide binding state of Gtr1, the different alleles of Gtr1 were expressed, together with Gtr2, in the presence or absence of Ego2. The interaction between Gtr1 and Gtr2 was indeed reduced in Ego2 WT cells expressing Gtr1S20L(Figure 2E), indicating that the nucleotide-binding state of Gtr1 is important for optimal dimerization in vivo. However, in the absence of Ego2, expression of GTP-locked Gtr1Q65L did not improve its interaction with Gtr2. Re-introduction of Ego2 in the same strain rescued the interaction of Gtr2 with both Gtr1 and Gtr1Q65L (Supplementary information, Figure S1). Hence, the stability of the Gtr1-Gtr2 heterodimer in vivo requires not only the binding of Gtr1 to GTP, but also its interaction with an intact EGOC at the vacuolar membrane.
Ego2 is essential for the assembly of the EGOC at the vacuolar membrane. (A) Subcellular localization of endogenously expressed Ego1-GFP and Ego3-GFP in the indicated deletion strains. Ego1-GFP localizes to the vacuolar membrane even in the absence of Ego2, Ego3, Ego4, or Ego2/Ego3 combined. Ego3-GFP requires both Ego1 and Ego2 to localize to the membrane. The lipophilic styryl dye FM4-64 was used to stain specifically the vacuolar membrane. (B) TAP pull-down experiments showing that the interaction between Ego1-TAP expressed from a plasmid under the control of its endogenous promoter and endogenously expressed Ego3-GFP is dependent on Ego2. Inputs (lysates of cells with the indicated genotypes) and TAP pull-down (eluted proteins after incubation with IgG sepharose) fractions were analyzed by immunoblotting with anti-TAP and anti-GFP antibodies. Ego1-GST expressing cells were used as a control. (C) Localization of Gtr1-GFP and Gtr2-GFP fusion proteins expressed from their endogenous promoters in the indicated deletion strains. Both Gtr1-GFP and Gtr2-GFP, and their activated versions, are mislocalized in the absence of Ego2. The vacuolar membranes were stained as in A. (D) TAP pull-down experiments showing that the interaction between Gtr1-TAP and Gtr2-V5 is reduced in ego1Δ (1Δ), ego2Δ (2Δ), and ego3Δ (3Δ), but not in ego4Δ (4Δ) mutant cells. (E) TAP pull-down experiments indicating that the reduced interaction between Gtr1-TAP and Gtr2-V5 in an ego2Δ strain is not rescued by expressing the GTP-locked version of Gtr1-TAP. In D and E lysates from cells expressing the indicated fusion proteins (input) and TAP pull-down fractions were analyzed by immunoblotting with anti-TAP and anti-V5 antibodies. Cells expressing untagged Gtr1 were used as a control.
Ego2 is a subunit of the EGOC
The structural homologs of Ego2 (and/or Ego4), LAMTOR4-LAMTOR5, interact with LAMTOR1, LAMTOR2, and LAMTOR3 in mammalian cells to complete the pentameric Ragulator complex at lysosomal membranes9. Since the loss of Ego2 had a clear detrimental effect on the integrity of the EGOC, we sought to determine whether Ego2 is also a subunit of the EGOC. To probe for potential interactions between Ego2 and the known EGOC components, we employed the split-ubiquitin yeast 2-hybrid system using either Ego2 or Ego4 as bait. Ego2 interacted most strongly with Ego1, but also with Ego3 and GTP-locked Gtr1Q65L (Figure 3A). Surprisingly, Ego4 also interacted with Gtr1 (WT and GTP-locked forms) and with Gtr2 (WT form). In this system, the bait protein is artificially localized to membranes. Since Ego2 and Ego4 are predicted to have highly similar structures23, it is possible that Ego4 interacts with Gtr1 and Gtr2 only when it is forced to localize to membranes, but not necessarily under physiological conditions. Consistent with such an explanation, Ego2-HA3 was found to co-precipitate with both Ego1-GFP and Ego3-GFP, whereas Ego4-HA3 showed no interaction with either protein (Figure 3B). To understand whether Ego2 and/or Ego4 could directly interact with the EGOC, we co-expressed codon-optimized versions of EGO2, EGO3, and EGO4 together with N-terminally His6-tagged EGO1 in bacteria, and performed a Ni-NTA affinity purification. Ego2 and Ego3, but not Ego4, co-purified in one complex with recombinant Ego1 (Figure 3C and 3D). This result was further confirmed by gel-filtration and GST pull-down assays (Supplementary information, Figure S2A-S2C). Thus, Ego2 can directly interact with Ego1-Ego3 to form a complex in vitro. Finally, we assessed the subcellular localization of Ego2-GFP and Ego4-GFP expressed under the control of their endogenous promoters. Fully consistent with our in vivo and in vitro analyses, the Ego2-GFP signal was enriched at the vacuolar periphery, whereas Ego4-GFP was observed uniquely as a diffuse cytosolic signal (Figure 3E). Like for Ego3 and the Gtr GTPases, the enrichment of Ego2-GFP at the vacuole was dependent on the presence of Ego1 (Figure 3F). The absence of Ego3 on the other hand did not affect the localization of Ego2-GFP and yeast 2-hybrid analysis performed in an ego3Δ strain confirmed that the interaction between Ego2 and Ego1 was indeed Ego3-independent (Figure 3G). Collectively, these data demonstrate that Ego2 physically associates with the EGOC at the vacuolar membrane through direct interactions with Ego1 and Ego3, strongly suggesting that Ego2 is a subunit of the EGOC that had previously escaped detection.
Ego2 interacts with Ego1 and Ego3 at the vacuolar membrane. (A) Interactions between Ego2 or Ego4 (baits) and the indicated EGOC components (preys) were assessed using the split ubiquitin-based membrane two-hybrid system (Dualsystems Biotech). Bait proteins were expressed from the pCAB vector as Cub-LexA-DBD fusion proteins and prey proteins from the pPR3-N vector as NubG-HA fusions in a strain containing the LexAop-HIS3, LexAop-ADE2, and LexAop-LacU reporter genes (NMY51). For each combination tested, β-galactosidase activity is expressed in Miller units as mean ± SD from at least three independent transformants grown overnight at 30 oC. pDL2-Alg5 and pAI-Alg5-NubI were used as negative and positive control preys, respectively. Values that were at least 15-fold higher than the negative control are shown in bold. (B) Ego2-HA3 co-immunoprecipitates with Ego1-GFP and Ego3-GFP. Lysates from ego2Δ or ego4Δ cells expressing the corresponding Ego2-HA3 or Ego4-HA3 fusions from plasmids under the control of their endogenous promoters together with endogenously expressed Ego1-GFP or Ego3-GFP (input) and HA-IP fractions were analyzed by immunoblotting with anti-HA and anti-GFP antibodies. ego2Δ or ego4Δ cells expressing either untagged Ego2 or Ego4 from plasmids were analyzed as controls. (C, D) His6-Ego1, Ego2, Ego3, and Ego4 were expressed in bacteria and subsequently purified through nickel affinity column followed by size-exclusion chromatography. Samples were taken at different steps and analyzed by SDS-PAGE followed by Coomassie blue staining (C) or immunoblot analysis (D) using antibodies raised against the indicated proteins. M, marker; N.I., not induced sample; I, IPTG-induced sample; P, purified sample. (E) Ego2-GFP, but not Ego4-GFP, is enriched at the yeast vacuolar periphery. Cells expressing Ego2-GFP or Ego4-GFP from a plasmid under the control of their endogenous promoters were visualized by microscopy in log phase. (F) Localizations of Ego2-GFP in the indicated deletion strains show that its vacuolar localization is dependent on the presence of Ego1. The vacuolar membranes were stained with FM4-64 in E and F. (G) Interactions between Ego2 (bait) and EGOC components (preys) were assessed using the split ubiquitin-based membrane two-hybrid system (Dualsystems Biotech) in NMY51 deleted for EGO3 and assessed as in A. The values indicate the mean β-galactosidase activity expressed in Miller units from at least three independent transformants grown overnight at 30 oC. Values that were at least 15-fold higher than the negative control are shown in bold.
Overall structure of the Ego1-Ego2-Ego3 ternary complex (EGO-TC)
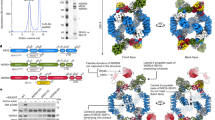
Our in vitro and in vivo assay results above show that Ego1, Ego2 and Ego3 can form a stable ternary complex (EGO-TC). To investigate the molecular basis of the biological function of the EGO-TC and the structure-function relationship of the component proteins, we carried out structural studies of the EGO-TC. We expressed an N-terminally truncated Ego1 (residues 17-184), full-length Ego2 (residues 1-75) and full-length Ego3 (residues 1-162) individually, and assembled the EGO-TC via co-purification by Ni-NTA affinity chromatography and gel-filtration chromatography. The crystal structure of the EGO-TC was determined at 2.4 Å resolution. In the crystal structure, there is one EGO-TC in an asymmetric unit, and the final structure model consists of residues 146-184 of Ego1, residues 8-73 of Ego2, and residues 2-162 of Ego3 (Figure 4A). There are no extensive interactions between symmetry-related molecules of Ego1, Ego2 and Ego3, excluding the possibility of dimerization.
Crystal structure of the EGO-TC. (A) Cylindrical cartoon presentation of the Ego1-Ego2-Ego3 complex with Ego1 in pink, Ego2 in yellow and Ego3 in green. (B-D) Detailed views of the interactions between Ego2 and Ego3 (B), between Ego1 and Ego2 (C), and between Ego1 and Ego3 (D). For clarity, residues of Ego1, Ego2, and Ego3 are labeled with superscripts 1, 2, and 3, respectively. Hydrogen bonds are indicated with dashed lines. (E) Sequence alignment of the C-terminal region of Ego1 from different species. The secondary structure of S. cerevisiaeEgo1 is placed above the alignment. Residues involved in the interactions with Ego2 and Ego3 are indicated with triangles. The sequence alignment was performed using the program ESPript37.
In the EGO-TC structure, only the C-terminal region (CTR, residues 146-184) of Ego1 is clearly defined although a nearly full-length Ego1 was present in the EGO-TC used in the crystallization. SDS-PAGE analysis of the crystallization solution showed that the N-terminal region (NTR, residues 17-145) of Ego1 was degraded during crystallization (Supplementary information, Figure S2D), suggesting that the NTR of Ego1 has no interactions with Ego2 and/or Ego3. The defined C-terminal region of Ego1 forms an α-helix (residues 154-172) flanked by an N-terminal loop (residues 146-155) and a C-terminal loop (residues 173-184); the α-helix is largely involved in interactions with Ego2 and the C-terminal loop largely in interactions with Ego3 (Figure 4A). Ego2 is well defined except the N-terminal residues 1-7 and C-terminal residues 74-75. Ego2 adopts a Roadblock domain fold consisting of a central core β-sheet of five anti-parallel β-strands (β2-β1-β5-β4-β3) flanked by a long α-helix on the bottom side, which is similar to LAMTOR5 (PDB code: 3MS6)11 with a root-mean-square deviation (RMSD) of 1.9 Å for 61 Cα atoms (Supplementary information, Figure S3A). Residues 2-162 of Ego3 are well defined except two surface-exposed loops (residues 52-63 and residues 88-92). Ego3 assumes a similar overall structure to our previously reported Ego3 structure (PDB code: 4FTX;20) with a RMSD of 0.7 Å for 112 Cα atoms (monomer), which is also similar to LAMTOR2 and LAMTOR312,13 (Supplementary information, Figure S3B and S3C). Intriguingly, our previous structural studies showed that Ego3 alone exists as a homodimer with a swapping architecture in which the N-terminal helix α1 of one monomer stretches over to the other monomer via a long α1-β1 loop20. However, in the EGO-TC, Ego3 exists as a monomer and Ego2 occupies the spatial position of the other monomer in the Ego3 homodimer (Supplementary information, Figure S3D). In addition, the N-terminal helix α1 folds along the core β-sheet to take the position of the swapping segment, and the long α1-β1 loop assumes a β-hairpin (named as the α1/β1 hairpin) to interact with Ego2 (Figure 4A).
Interactions among different Ego proteins
Ego1, Ego2, and Ego3 form a tight ternary complex and the interaction interfaces bury 4 388 Å2 (or 36.2%) of the total solvent-accessible surface area of 11 979 Å2 as analyzed by the PISA program25. Ego2 and Ego3 form a heterodimer that is flanked along one side by Ego1 (Figure 4A). The interaction interface between Ego2 and Ego3 buries a total of 931 Å2 solvent-accessible surface areas, which is much less than that of the Ego3 homodimer (4 140 Å2) and those of the Ego1-Ego2 (1 493 Å2) and Ego1-Ego3 (2 131 Å2) interfaces. At the Ego2-Ego3 interface, Tyr96 of Ego3 makes two hydrogen bonds with Asp16 and Asn20 and a π-π stacking interaction with His18 of Ego2 via their side chains (Figure 4B). In addition, the main-chain carbonyl of Val21 of Ego2 forms two hydrogen bonds with the main-chain amides of Gly140 and Leu141 of Ego3. Val25, Phe27, Pro138, and Leu141 of Ego3 and Thr12, Val21, and Glu23 of Ego2 are involved in hydrophobic interactions. The interaction interface between Ego2 and Ego3 is significantly different from the common side-by-side mode as observed in the structures of the Ego3 homodimer and many other Roadblock domain-containing dimeric proteins such as the heterodimeric LAMTOR2-LAMTOR3, or the homodimeric LAMTOR5 and MglB11,12,13,26. For these proteins, the interaction interface is usually very large and involves the dimerization of the core β-sheet to form an extended 10- or 12-stranded β-meander, as well as extensive interactions between the long α-helices on the bottom side. In the structure of the EGO-TC, the β-sheets of Ego2 and Ego3 are in different orientations and cannot form an extended β-sheet, and the two α-helices of Ego2 and Ego3 on the bottom side are nearly 30 Å apart (Supplementary information, Figure S3D). These results together indicate that Ego2 and Ego3 interact relatively weakly with each other in the absence of Ego1, which is consistent with the GST pull-down assays (Supplementary information, Figure S2B and S2C).
In the EGO-TC, the N-terminal part of the α-helix of Ego1 lies along the upper side, and the N-terminal loop folds along one edge of the core β-sheet of Ego2 (Figure 4C). The interactions between Ego1 and Ego2, which bury a total of 1 493 Å2 solvent-accessible surface areas, are largely hydrophobic and involve hydrophobic residues Ile152, Leu157, Leu160, His161, Ile164, Leu165, and Ile168 of Ego1 and Ile22, Phe48, His59, Tyr61, Ile66, Leu68, and Val70 of Ego2. In addition, the main chain of Pro147 of Ego1 forms a hydrogen bond with the side chain of Gln50 of Ego2, and the side chain of His161 of Ego1 forms two hydrogen bonds with the main chains of both Arg63 and Ile66.
The C-terminal part of the α-helix of Ego1 has extensive hydrophilic and hydrophobic interactions with the α1/β1 hairpin and helix α3 of Ego3, and the C-terminal loop of Ego1 interacts with helices α1 and α3 of Ego3 (Figure 4D). The interactions between Ego1 and Ego3 bury a total of 2 131 Å2 solvent-accessible surface areas. Specifically, the side chain of Glu167 of Ego1 forms five hydrogen bonds with His23 and Ser24 of Ego3; the side chain of Gln171 of Ego1 forms two hydrogen bonds with the main chain of Asp21 of Ego3, and the side chain of Ser172 forms a hydrogen bond with the side chain of Tyr20 of Ego3. In addition, the α-helix of Ego1 is further stabilized by numerous hydrophobic interactions among Leu160, Ile164, and Ile168 of Ego1 and Tyr20, and Val25 of the α1/β1 hairpin, and Leu141 and Ile144 of helix α3 of Ego3. The C-terminal loop of Ego1 lies in a surface cleft formed by helices α1 and α3 of Ego3. The main chain of Asn175 of Ego1 forms a hydrogen bond with the side chain of Asn15 of Ego3; the side chain of Lys176 of Ego1 makes a salt-bridge interaction with the side chain of Asp155; the main chain of Leu180 of Ego1 makes a hydrogen bond with the side chain of His5 of Ego3, and the main chain of Val182 of Ego1 makes two hydrogen bonds with the main chain of Met3 of Ego3. In addition, Val174, Leu180, and Ile182 of Ego1 make numerous hydrophobic contacts with Met3, Val9, Phe12, and Leu153 of Ego3. Interestingly, sequence alignment of the C-terminal region of Ego1 from different species shows that most of the residues involved in the interactions with Ego2 and Ego3 are strictly or highly conserved (Figure 4E), suggesting that these interaction interfaces are very likely conserved in other species.
Functional validation of the interactions between different Ego proteins
The interactions of Ego1 with Ego2 and Ego3 were validated using the in vitro GST pull-down assay and in vivo split-ubiquitin yeast 2-hybrid system. Consistent with our structure, we found that Ego2 can form a complex with Ego1 and the C-terminally truncated Ego1Δ169-184 variant, whereas Ego2 can interact with Ego3 only in the presence of the full-length Ego1, but not the Ego1Δ169-184 variant (Figure 5A and Supplementary information, Figure S2B). Expansion of the C-terminal truncation in Ego1 to residue 152 abolished the interaction of the respective Ego1Δ152-184 variant with both Ego2 and Ego3. To investigate the functional importance of the different regions, we assessed the ability of each truncation to rescue the EGOC localization defects observed in ego1Δ cells. For these analyses, WT Ego1 and the two truncated forms (Ego1Δ169-184 and Ego1Δ152-184) were fused with a HA3 tag at their C-terminus, allowing us to verify that all forms were expressed to similar amounts in yeast. In agreement with our structural data, the vacuolar localization of Ego2-GFP, Ego3-GFP, Gtr1-GFP, and Gtr2-GFP was abolished in strains expressing Ego1Δ152-184 (Figure 5B). The residues in Ego1 spanning from position 169-184 are also essential for the vacuolar recruitment of Ego3 and Gtr GTPases. Although our in vitro and 2-hybrid data imply that the interaction between Ego2 and Ego1 does not require residues 169-184 of Ego1, we observed only a weak Ego2-GFP signal at the vacuole in Ego1Δ169-184-expressing cells. It is possible, however, that the GFP tag interferes with the formation of all the necessary interactions between Ego1 and Ego2. Though both the HA3-tagged and untagged versions of WT Ego1 could rescue the rapamycin sensitivity of ego1Δ cells, expression of either of the truncated Ego1 forms resulted in a severe growth defect in the presence of rapamycin equal to that of cells lacking Ego1 completely (Figure 5C). Moreover, compared to Ego1, both Ego1 truncated variants led to reduced TORC1 activity as assessed by the extent of Sch9 phosphorylation (Figure 5D), indicating that this entire C-terminal stretch is necessary for Ego1 function in TORC1 signaling. Collectively, these data indicate that the residues of Ego1 involved in binding both Ego2 (i.e., residues 152-168 in Ego1) and Ego3 (i.e., residues 169-182 in Ego1) are essential and equally important for the function of Ego1 in TORC1 activation.
Functional analyses of the truncated Ego1 variants. (A) Interactions between Ego2 or Ego3 (baits) and the Ego1 variants (preys) were assessed using the split ubiquitin-based membrane two-hybrid system (Dualsystems Biotech). For each combination tested, cells were serially (10-fold) diluted and spotted onto SD-LT (control) or SD-LTA medium and β-galactosidase activity was measured. β-galactosidase activity is expressed in Miller units as mean ± SD from at least three independent transformants grown overnight at 30 °C. Empty bait vector pCabWT and pDl2-Alg5 prey were used as negative controls. Values that were at least 15-fold higher than the negative control prey are shown in bold. The full-length Ego1 interacts with both Ego2 and Ego3, whereas the truncated Ego1Δ169-184 interacts with Ego2, but only very weakly with Ego3. The truncated Ego1Δ152-184 interacts with neither Ego2 nor Ego3. (B) Ego2-GFP, Ego3-GFP, Gtr1-GFP, and Gtr2-GFP localizations in ego1Δ cells expressing an empty vector or WT Ego1-HA3, Ego1Δ152-184-HA3, or Ego1Δ169-184-HA3 expressed from plasmids under the control of their own promoter. (C) The rapamycin sensitivity of an ego1Δ strain cannot be rescued by expression of Ego1Δ152-184 or Ego1Δ169-184. Serial 10-fold dilutions of the indicated strains were spotted onto YPD plates containing 0, 4, or 6 ng/ml rapamycin. (D) Immunoblots detecting the extent of phosphorylation within the C-terminus of Sch9 were used to quantify TORC1 activity (the ratio of hyperphosphorylated (+P)/hypophosphorylated (−P) Sch9) in the indicated strains. The values (means ± SD; n = 3) were normalized to WT cells and presented in the bar graph. One representative immunoblot is shown.
We also performed mutagenesis studies to validate the interactions of Ego2 with Ego1 and Ego3. Our in vitro GST pull-down assay showed that mutations of two representative residues of Ego2 on the Ego2-Ego1 interface (I66D and L68D) could disrupt its interaction with Ego1 and consequently with Ego3 (Supplementary information, Figure S2C). However, a triple mutation of Ego2 on the Ego2-Ego3 interface (D16A/H18A/N20A) had no significant effect on the assembly of the EGO-TC (Supplementary information, Figure S2C). These results are consistent with the structural data showing that the interaction between Ego2 and Ego3 is relatively weak and is mainly mediated by Ego1.
In our previous studies of Ego3, we performed a series of mutagenesis and in vivo functional assays to evaluate the functional roles of the α1/β1 loop and several conserved residues including Tyr96 in strand β3 of Ego3 on the dimer interface20. Based on the structural and functional data, we suggested that the dimerization of Ego3 is essential for the function of the EGOC. However, a detailed analysis of those mutagenesis data show that most of the residues involved in the dimer interface and the N-terminal α-helix are also involved in the interactions with Ego1 and Ego2, and the previous mutagenesis and functional data could be better explained in the context of the EGO-TC. Specifically, the Ego3 M1 mutant contains a shortened α1/β1 loop (KPYDLHSVDFKTSSL was replaced by KPYGSL), and thus would impair or disrupt the interaction of Ego3 with Ego1 and consequently failed to accumulate at the vacuolar periphery. Consistent with the triple mutation of Ego2 (Supplementary information, Figure S2C), mutation Y96E of Ego3 on the interaction interface with Ego2 would not impair the assembly of the EGO-TC, explaining why the Ego3 M2 mutant containing the Y96E mutation had no significant impact on Ego3-GFP localization or rapamycin-sensitivity. The Ego3 M3 mutant (M1 and M2 mutants) would further impair or disrupt the interactions of Ego3 with Ego1 and Ego2 and prevent the formation of the EGO-TC, and thus the recruitment of Gtr1-Gtr2 to the vacuolar periphery.
The EGO-TC functions as a scaffold for the Gtr GTPases at the vacuolar membrane
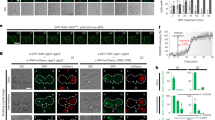
While our data clearly demonstrate that the EGO-TC is essential for recruiting the Gtr1-Gtr2 heterodimer to the vacuolar membrane, it remained unclear whether the EGO-TC might also have an additional function in Gtr activation analogous to the proposed GEF activity of the Ragulator9. We therefore asked whether artificially tethering Gtr1-Gtr2 to the vacuolar membrane would be sufficient to promote TORC1 activity in the absence of the EGO-TC. To this end, we fused the first 151 amino acids of Ego1 (which when expressed alone is not functional in TORC1 signaling; Figure 5) to vhhGFP4, a single-domain antibody fragment directed against GFP (Ego1NT-vhhGFP)27,28,29. In cells lacking the EGO-TC, Gtr1-GFP was mislocalized in the cytosol. However, in EGO-TC-deleted cells also expressing the Ego1NT-vhhGFP4 construct, we observed a clear Gtr1-GFP signal at the vacuolar periphery (Figure 6A). The presence of the Ego1NT-vhhGFP4 construct also rescued both the rapamycin recovery defect and low TORC1 activity of cells lacking the EGO-TC (Figure 6B and 6C). Furthermore, TORC1 remained responsive to leucine starvation and re-addition, demonstrating that vacuolar-tethered Gtr1-Gtr2 are still subject to amino acid regulation in the absence of the EGO-TC (Figure 6D). Gtr1-GFP tethered to the vacuolar membrane via a Vac8-vhhGFP4 construct also remained able to regulate TORC1 activity in a leucine-dependent manner, excluding the possibility that the N-terminus of Ego1 itself is sufficient to activate the Gtr-dependent TORC1 signaling (Figure 6E). These data suggest that the principal function of the EGO-TC is to provide a scaffold for the Gtr1-Gtr2 heterodimer at the vacuolar membrane.
Vacuolar-tethered Gtr1-Gtr2 is sufficient to promote the amino acid-dependent TORC1 activity in the absence of the EGO-TC. (A) Ego1NT-vhhGFP4 targets Gtr1-GFP to the vacuolar membrane in the absence of the EGO-TC. Gtr1-GFP and Gtr2 were expressed from plasmids in gtr1Δ gtr2Δ or egocΔ (ego1Δ ego2Δ ego3Δ gtr1Δ gtr2Δ) deleted cells in the absence or presence of Ego1NT-vhhGFP4. (B) The rapamycin recovery defect of a strain lacking the EGO-TC (and expressing Gtr1-GFP) is rescued in the presence of Ego1NT-vhhGFP4. The indicated strains were treated with 200 ng/ml rapamycin for 6 h in liquid medium before serial 10-fold dilutions were spotted onto solid YPD medium. (C) The low TORC1 activity in EGO-TC-deleted, Gtr1-GFP expressing cells is rescued in the presence of Ego1NT-vhhGFP. Immunoblots detecting the extent of phosphorylation within the C-terminus of Sch9 were used to quantify TORC1 activity (the ratio of hyperphosphorylated (+P)/hypophosphorylated (−P) Sch9) in the indicated strains. The values (means ± SD; n = 3) were normalized to WT cells and presented in the bar graph. One representative immunoblot is shown. (D, E) TORC1 activity is regulated in a leucine-dependent manner in EGO-TC-deleted cells when Gtr1-GFP is tethered to the vacuolar membrane via either (D) Ego1NT-vhhGFP4 or (E) Vac8-vhhGFP4. The indicated strains were made auxotrophic for leucine and grown to exponential phase in leucine-containing medium. Cells were then filtered, washed twice, and starved for leucine for up to 60 min (− Leucine) before re-addition of leucine (2.8 mM final concentration) for up to 15 min (+ Leucine). Samples were taken at the indicated time points and TORC1 activity was assayed as in C.
Discussion
Biological implication of the Ego1-Ego2-Ego3 complex
In the present study, we show that the previously uncharacterized protein Ego2 is an additional and essential component of the EGOC. Our structural and functional data demonstrate that Ego2 interacts directly with Ego1 and Ego3 to form a stable complex, of which all three components are necessary for the vacuolar recruitment of the Gtr1-Gtr2 GTPases and hence the activation of TORC1 signaling in response to the presence of amino acids.
The previous structural studies have shown that Ego3 alone exists as a homodimer both in solution, as shown by gel-filtration analysis (Supplementary information, Figure S2A), and in the crystal structure20, where it displays a swapping conformation and a dimer interface larger than its interaction interfaces with Ego1 and Ego2 in the EGO-TC (see Results above). It has also been shown that at low pH, Ego3 alone can form a tetramer in which the four Ego3 monomers are assembled in an X-shape manner linked together by the swapping α1 helix and the α1/β1 β-strand, instead of forming an extended, inter-molecular β-sheet as observed in the Ego3 homodimer19. Consequently the monomer-monomer interface is much looser than that of the Ego3 homodimer. However, in the EGO-TC, Ego1, Ego2, and Ego3 form a stable 1:1:1 TC without dimerization and the site at which Ego3 would interact with another monomer of Ego3 in the homodimer or tetramer is occupied by Ego2 (Figure 4A). Interestingly, structural studies show that, while LAMTOR5 alone exists as a homodimer11, it can also form a heterodimer with LAMTOR4 in which its LAMTOR4 binding site largely overlaps with the dimer interface in its homodimeric form (Zhang et al., unpublished results). These results indicate that the Ego3 monomer, rather than the Ego3 homodimer or tetramer, interacts with Ego1 and Ego2, and formation of the Ego3 homodimer would in fact prevent the binding of Ego3 with Ego1 and Ego2, and hence the assembly of the EGO-TC. There are two possibilities to assemble this complex. In the cytoplasm at neutral pH condition, most of Ego3 might exist mainly as a homodimer. However, at the membranes, which are enriched with acidic phospholipids and have a lower pH value (1.6 unit) than that in the cytoplasm30, the Ego3 homodimer might be partially dissociated allowing Ego3 to adopt an intermediate conformation similar to the Ego3 tetramer, which could be further dissociated and then interact with Ego1 and Ego2 to form the EGO-TC.
On the other hand, we were able to express the Ego proteins separately and then assemble them together to form the EGO-TC in vitro at a neutral pH value (pH 7.5; see Materials and Methods). More detailed analyses showed that, although our in vitro and in vivo data indicate that Ego3 alone could not interact with Ego2 to form a binary complex, Ego3 could interact weakly with Ego1 in vitro as shown by the previous gel-filtration analysis19 and our GST pull-down assay (Supplementary information, Figure S2B). In addition, in the structures of both the Ego3 homodimer and the tetramer, helix α3 and the swapping helix α1 form a cleft similar to the binding site of Ego3 for Ego1 in the EGO-TC. Based on these results, it is also possible that in the cytoplasm, Ego3 exists mainly as a homodimer and cannot interact tightly with Ego1 or/and Ego2 to activate TORC1 signaling. Due to the lack of knowledge about the presence of Ego2, we previously suggested that Ego3 functioned as a homodimer to mediate the interaction between Ego1 and Gtr1-Gtr2 in the EGOC to activate TORC120. It seems more likely that in the cytoplasm Ego3 alone might exist mainly as a homodimer to avoid mis-activating TOR signaling; however, when it is recruited to the vacuolar membrane by Ego1-Ego2, the interaction between Ego3 and Ego1-Ego2 may facilitate the conformational changes of Ego3 from the homodimer to the monomer, resulting in the formation of the EGO-TC, which can further promote the vacuolar membrane localization of Gtr1-Gtr2 and then activate TOR signaling.
Comparisons of the Ego and LAMTOR proteins
The previous studies have shown that the yeast Ego proteins are the equivalents of the LAMTOR proteins in the mammalian Ragulator complex14 (Figure 7A). Interestingly, Ego2-4 and LAMTOR2-5 proteins all contain a Roadblock domain which consists of a common α-β-α sandwich fold with the central β-sheet flanked by one α-helix on the bottom side and one or two α-helices on the upper side23. Although the Roadblock domains in these proteins are structurally conserved, there are notable differences9,21. Based on their structural features we can divide them into two groups: group I contains Ego2, Ego4, LAMTOR4, and LAMTOR5, and group II includes Ego3, LAMTOR2, LAMTOR3, and the C-terminal Roadblock domains of Rag and Gtr GTPases (Figure 7B). Both LAMTOR5 (PDB code: 3MSH) and Ego4 (PDB code: 2GRG) exhibit a fold of αββαββ. In the EGO-TC, Ego2 adopts a fold of ββαββ, which lacks the N-terminal α-helix compared with Ego4 and LAMTOR5. LAMTOR4 was predicted to share a similar fold as LAMTOR5 and Ego29. The main feature of those domains is that there is no or only one short α-helix (α1) on the upper side of the central β-sheet. Most of the group II proteins share a common fold of αββαβββα with an additional C-terminal α-helix (α3) on the upper side compared with the group I proteins. In particular, Ego3 has an extra β-hairpin and β-strand and assumes a fold of αβββαββββα.
Comparison of Ego proteins and LAMTOR proteins. (A) Secondary structure comparison of Ego1-3 and LAMTOR1-5 based on the crystal structures, except for Ego1, LAMTOR1 and LAMTOR4, whose secondary structures were predicted using the PredictProtein server38. (B) Structural comparison of Ego2, Ego3, Ego4, LAMTOR2, LAMTOR3, and LAMTOR5. These Roadblock domain proteins can be divided into two groups: group I contains Ego2, Ego4, and LAMTOR5, and group II contains Ego3, LAMTOR2, and LAMTOR3. The potential binding sites for protein partners are marked with oval frames.
Due to the lack of a long α-helix on the upper side of the β-sheet, the group I proteins contain a mortise-liked surface which could act as a potential binding site for other protein partner(s). For example, as a typical group I protein, Drosophila melanogaster LC7 uses the flat surface on the upper side of the β-sheet to interact with the N-terminal domain of dynein intermediate chain (IC)31 (Supplementary information, Figure S3E). Specifically, one helix of the IC lies on the upper side of LC7 to form a helix bundle with the N-terminal helix of LC7. In the EGO-TC, Ego2 also has a binding site for Ego1 on the upper side of the β-sheet. It is worth noting that Ego2 lacks an N-terminal helix compared with the other group I proteins. Thus, Ego2 could not stabilize the binding segment of Ego1 by itself (Figure 7 and Supplementary information, Figure S3E). Interestingly, in the EGO-TC, the α1/β1 hairpin of Ego3 occupies the position of the missing N-terminal 1 helix of other group I proteins and assumes its functional role in stabilizing the -helix of Ego1 through multiple hydrogen-bonding and hydrophobic interactions (Supplementary information, Figure S3E). In analogy to these structural arrangements in yeast, it is tempting to predict that LAMTOR1 is likely to interact via a long α-helix with the flat surface on the upper side of the β-sheets of the LAMTOR4-LAMTOR5 complex in humans.
As the group II proteins have two helices on the upper side, there is not enough space to accommodate another helix from a protein partner. On the other hand, there is a surface cleft formed by helices α1 and α3 of Ego3 which interacts with the C-terminal loop of Ego1. It is possible that the cleft between the two helices (α1 and α3) in other group II proteins could be a potential binding site for a protein partner. For example, the corresponding clefts on the LAMTOR2 and LAMTOR3 might be the binding site for LAMTOR1 (Figure 7B).
In conclusion, the human Ragulator complex is structurally and functionally the counterpart of the yeast EGO-TC complex. Although the mammalian Ragulator comprises more components than the EGO-TC, based on the structural similarities between the component proteins it is tempting to speculate that the Ragulator is assembled in a similar manner. LAMTOR1 may act as a scaffold analogous to Ego1 and directly interact with the LAMTOR2-LAMTOR3 and LAMTOR4-LAMTOR5 heterodimers, which would assume the positions of Ego3 and Ego2, respectively. Surprisingly, whereas the pentameric Ragulator complex has been described to promote the GTP-bound state of RagA/B in vitro, our analysis of EGO-TC function in vivo suggests that the principal role of the EGO-TC is to provide a scaffold for the Gtr1-Gtr2 heterodimer at the vacuolar membrane. It is possible that another protein or proteins may provide GEF activity toward Gtr1 in the presence of amino acids; for example, Vam6 was shown to exhibit a GEF activity toward Gtr114. It is also possible that the EGOC might contain other components and the whole complex may possess a GEF activity toward Gtr1. To this end, the NTR of Ego1 not seen in the EGO-TC structure might provide binding site(s) for additional component(s). Structural and functional studies are currently ongoing to further elucidate the mechanism(s) by which the Gtr1-Gtr2 GTPases are regulated in response to amino acids.
Materials and Methods
Growth condition
All strains and plasmids used in this work are listed in Supplementary information, Tables S1 and S2, respectively. Unless otherwise stated, all strains were made prototrophic with the appropriate plasmids before use. For each experiment, cells were pre-grown overnight in synthetic dropout (SD) medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 0.2% dropout mix, and 2% glucose), then diluted to an OD600 of 0.2 and further grown at 30 °C until they reached exponential growth (OD600 of 0.8). Expression of genes under the control of the TetON promoter was induced by adding 5 μg/ml doxycycline to the medium.
Sch9 phosphorylation analysis
Sch9T570A-HA5 C-terminal phosphorylation was monitored following 2-nitro-5-thiocyanatobenzoic acid (NTCB)-induced chemical cleavage as described previously24. Quantification of TORC1 activity (extent of Sch9 phosphorylation) was determined as described previously22. Briefly, NTCB was added to lysed cell extracts to induce chemical cleavage. Extracts were separated on an SDS-PAGE gel and membranes were probed with anti-HA antibodies (12CA5) and anti-mouse HRP-conjugated antibodies (Bio-Rad).
Co-immunoprecipitation
Yeast cells expressing the indicated fusion proteins were harvested by filtration and stored at −80 °C. Cells were resuspended in a lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.2% NP40, and protease and phosphatase inhibitor cocktails; Roche) and lysed with glass beads using the Precellys cell disruptor. For input samples, lysates were concentrated by precipitation with ice-cold acetone, resuspended in 6× concentrated loading buffer and denatured for 10 min at 95 °C. For co-immunoprecipitation, lysates were incubated for 2 h at 4 °C with prewashed IgG Sepharose beads (GE Healthcare) for TAP pull-downs or anti-HA magnetic beads (Pierce) for HA immunoprecipitation. Beads were washed three times with the lysis buffer before resuspension in 6× concentrated loading buffer and denaturation for 10 min at 95 °C. Inputs and IP samples were analyzed by SDS-PAGE immunoblotting using anti-GFP (Roche), anti-HA (Roche), anti-TAP (Open Biosystems), and anti-V5 (Invitrogen) antibodies together with light-chain specific anti-mouse or anti-rabbit HRP-conjugated antibodies (Jackson Immuno Research).
Microscopic analysis
Mid-log phase cells cultured in SD medium were imaged using an Olympus BX51 microscope (Olympus) equipped with a piezo positioner (Olympus), a XBO 75 W Xenon light source (Atlanta Light Bulbs, GA, USA), 100× 1.45 Plan-Fluor objectives, and a three-position filter sliding rack containing the filter sets U-MWIBA, U-MWIG and U-MNUA2 (Olympus). Images were acquired with an F-view2 camera (Olympus). CellM software (Olympus) was used to obtain and analyze the images. Representative images from several independent experiments in which at least 50 cells were analyzed are shown as the results.
Split-ubiquitin yeast two-hybrid assay
The membrane-based yeast two-hybrid system provided by Dualsystems Biotech was used following the manufacturer's instructions. Briefly, the NMY51 strain containing the LexAop-HIS3, LexAop-ADE2, and LexAop-LacZ reporter genes was co-transformed with a bait plasmid (pCAB-WT) expressing a Cub-LexA-DBD fusion protein from the CYC1 promoter and a prey plasmid (pPR3-N) expressing a NubG-HA fusion protein from the CYC1 promoter. pDL2-Alg5 (mutated NubG) and pAI-Alg5-NubI (high affinity for Cub) were used as negative and positive control preys, respectively. Cells were grown in synthetic medium without leucine and tryptophan. Interactions were assessed by spotting serial dilutions of cells onto solid SD medium without leucine, tryptophan, and adenine and/or by performing classical β-galactosidase assays.
Cloning, expression, and purification of the Ego proteins
Codon-optimized versions of His6-EGO1, EGO2, EGO3, and EGO4 were co-expressed in E. coli RosettaTM cells after induction with 0.5 mM IPTG for 3 h at 30 °C. Cells were collected by centrifugation, resuspended in buffer A (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1.5 mM MgCl2, 5% glycerol, and 20 mM imidazole) and lysed with a microfluidizer. The lysate was passed through a 1 ml HisTrap FF crude column (GE Healthcare), and washed with the same buffer before elution with buffer A containing 500 mM imidazole. The sample was further purified through a 16/60 Superdex 200 column (GE Healthcare) equilibrated with buffer B (20 mM HEPES, pH 7.5, 150 mM NaCl, 10 mM KCl, and 10 mM MgCl2). The purified EGOC was concentrated by centrifugation with a Vivaspin 500 column (10 000 Dalton MWCO, Sartorius), glycerol was added to a final concentration of 20%, and proteins were snap frozen in liquid nitrogen and stored at −80 °C. Proteins were separated on an SDS-PAGE gel and stained with Coomassie blue or transferred to a nitrocellulose membrane and probed with antibodies raised in rabbit against Ego1, Ego2, Ego3, or Ego4 (GenScript) and anti-rabbit HRP-conjugated antibodies (Bio-Rad).
To obtain the Ego1-Ego2-Ego3 complex for structural studies, the genes encoding the N-terminal truncated Ego1 (residues 17-184) and the full-length Ego3 (residues 1-162) were separately inserted into the NcoI and XhoI restriction sites of pET-Duet vector (Novagen) without tags, and the gene encoding the full-length Ego2 (residues 1-75) was inserted into the BamHI and XhoI sites of a modified pET-28a vector that then mediated expression of an N-terminally His6-SUMO tagged Ego2 protein. The three plasmids were transformed individually into E. coli BL21 (DE3) Codon-Plus strain (Stratagene). The bacterial cells were grown at 37 °C in LB medium until OD600 reached 0.8, and then induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 20 °C for 20 h. The cells overexpressing the three proteins were mixed together and then lysed by sonication in a lysis buffer (30 mM Tris-HCl, pH 7.5, and 200 mM NaCl). The proteins were purified by affinity chromatography using a Ni-NTA column (Qiagen) with the lysis buffer supplemented with 30 mM imidazole and 200 mM imidazole serving as washing buffer and elution buffer, respectively. The N-terminal His6-SUMO tag of Ego2 in the purified Ego1-Ego2-Ego3 complex was removed by a ubiquitin-like protease 1 and the complex was further purified by gel filtration using a Superdex 75 10/300 column (preparative grade; GE Healthcare) pre-equilibrated with a storage buffer (10 mM HEPES, pH 7.5, 100 mM NaCl, 2 mM MgCl2, and 1 mM DTT). For the proteins used in pull-down assays, genes encoding the full-length Ego1, Ego2, Ego3, and the Ego1Δ169-184 and Ego1Δ152-184 variants were cloned into pET-28a vector that allows C-terminal fusion of a His6 tag. The full-length Ego2, Ego3, and Ego4 were cloned into the pET-M30 vector to obtain GST-fused proteins. The expression and purification of these proteins were performed as described previously20. The Ego1, Ego2, and Ego3 proteins alone and the Ego1-Ego2 and Ego1-Ego2-Ego3 complexes were of sufficient purity (> 95%) as analyzed by SDS-PAGE and of high homogeneity as shown by gel-filtration analyses (Supplementary information, Figure S2A). Expression and purification of the Ego1 and Ego2 mutants were the same as for the WT proteins.
Crystallization, data collection, and structure determination
Crystallization of the Ego1-Ego2-Ego3 complex was performed using the hanging-drop vapor-diffusion method by mixing 2 μl protein solution (about 10 mg/ml) and 2 μl reservoir solution at 16 °C. Crystals of the Ego1-Ego2-Ego3 complex were grown from drops consisting of a reservoir solution of 0.2 M NaCl, 0.1 M Bis-Tris (pH 5.5) and 25% w/v polyethylene glycol 3,350 (PEG3350) in about 2 months. A 2.4 Å resolution diffraction data set was collected at −175 °C at beamline 17U of Shanghai Synchrotron Radiation Facility, China, and was processed, integrated, and scaled together with HKL200032. The statistics of the diffraction data are summarized in Table 1. Initial phases were obtained by the molecular replacement method implemented in Phenix33 using Ego3 (PDB code: 4FTX) and LAMTOR5 (PDB code: 3MS6, which has high similarity with Ego2 in the length and secondary structures) as the search models. Structure refinement was carried out using Phenix and Refmac533,34. The stereochemistry of the structure model was analyzed using MolProbity35. Structure analysis was carried out using programs in CCP436. The structure figures were prepared using Pymol (http://www.pymol.org). The statistics of the structure refinement and the quality of the structure model are also summarized in Table 1.
Accession code
The coordinates and structure factors of the Ego1-Ego2-Ego3 complex have been deposited in the RCSB Protein Data Bank with accession code: 4XPM.
Accession codes
References
Shimobayashi M, Hall MN . Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol 2014; 15:155–162.
Laplante M, Sabatini DM . mTOR signaling in growth control and disease. Cell 2012; 149:274–293.
Jewell JL, Russell RC, Guan KL . Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 2013; 14:133–139.
Kim J, Guan KL . Amino acid signaling in TOR activation. Annu Rev Biochem 2011; 80:1001–1032.
Efeyan A, Zoncu R, Sabatini DM . Amino acids and mTORC1: from lysosomes to disease. Trends Mol Med 2012; 18:524–533.
Duran RV, Hall MN . Regulation of TOR by small GTPases. EMBO Rep 2012; 13:121–128.
Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008; 320:1496–1501.
Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL . Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 2008; 10:935–945.
Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM . Ragulator is a GEF for the Rag GTPases that signal amino acid levels to mTORC1. Cell 2012; 150:1196–1208.
Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM . Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010; 141:290–303.
Garcia-Saez I, Lacroix FB, Blot D, Gabel F, Skoufias DA . Structural characterization of HBXIP: the protein that interacts with the anti-apoptotic protein survivin and the oncogenic viral protein HBx. J Mol Biol 2011; 405:331–340.
Kurzbauer R, Teis D, de Araujo MEG, et al. Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen-activated protein kinase signaling to late endosomes. Proc Natl Acad Sci USA 2004; 101:10984–10989.
Lunin VV . The Structure of the MAPK Scaffold, MP1, Bound to its partner, p14: A complex with a critical role in endosomal MAP kinase signaling. J Biol Chem 2004; 279:23422–23430.
Binda M, Péli-Gulli MP, Bonfils G, et al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell 2009; 35:563–573.
Panchaud N, Péli-Gulli MP, De Virgilio C . SEACing the GAP that nEGOCiates TORC1 activation: evolutionary conservation of Rag GTPase regulation. Cell Cycle 2013; 12:2948–2952.
Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C . The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell 2005; 19:15–26.
Keppler-Ross S, Noffz C, Dean N . A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans. Genetics 2008; 179:705–710.
Ashrafi K, Farazi TA, Gordon JI . A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J Biol Chem 1998; 273:25864–25874.
Kogan K, Spear ED, Kaiser CA, Fass D . Structural conservation of components in the amino acid sensing branch of the TOR pathway in yeast and mammals. J Mol Biol 2010; 402:388–398.
Zhang T, Péli-Gulli MP, Yang H, De Virgilio C, Ding J . Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the EGO complex to activate TORC1. Structure 2012; 20:2151–2160.
Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C . Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell 2012; 46:105–110.
Panchaud N, Péli-Gulli MP, De Virgilio C . Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci Signal 2013; 6:ra42.
Levine TP, Daniels RD, Wong LH, Gatta AT, Gerondopoulos A, Barr FA . Discovery of new Longin and Roadblock domains that form platforms for small GTPases in Ragulator and TRAPP-II. Small GTPases 2013; 4:62–69.
Urban J, Soulard A, Huber A, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 2007; 26:663–674.
Krissinel E, Henrick K . Inference of macromolecular assemblies from crystalline state. J Mol Biol 2007; 372:774–797.
Miertzschke M, Koerner C, Vetter IR, et al. Structural analysis of the Ras-like G protein MglA and its cognate GAP MglB and implications for bacterial polarity. EMBO J 2011; 30:4185–4197.
Caussinus E, Kanca O, Affolter M . Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol 2012; 19:117–121.
Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H . A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics 2008; 7:282–289.
Rothbauer U, Zolghadr K, Tillib S, et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods 2006; 3:887–889.
van der Goot FG, Gonzalez-Manas JM, Lakey JH, Pattus F . A 'molten-globule' membrane-insertion intermediate of the pore-forming domain of colicin A. Nature 1991; 354:408–410.
Hall J, Song Y, Karplus PA, Barbar E . The crystal structure of dynein intermediate chain-light chain roadblock complex gives new insights into dynein assembly. J Biol Chem 2010; 285:22566–22575.
Otwinowski Z, Minor W . Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 1997; 276:307–326.
Adams PD, Afonine PV, Bunkóczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr 2010; 66:213–221.
Murshudov GN, Vagin AA, Dodson EJ . Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr 1997; D53:240–255.
Davis IW, Leaver-Fay A, Chen VB, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 2007; 35:W375–W383.
Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr 2011; D67:235–242.
Gouet P, Courcelle E, Stuart DI, Metoz F . ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 1999; 15:305–308.
Yachdav G, Kloppmann E, Kajan L, et al. PredictProtein-an open resource for online prediction of protein structural and functional features. Nucleic Acids Res 2014; 42:W337–W343.
Acknowledgements
We thank the staff members at BL17U of Shanghai Synchrotron Radiation Facility (SSRF), China for technical support in diffraction data collection, Riko Hatakeyama and Markus Affolter for plasmids, and Floriane Jaquier for technical assistance. This research was supported by the Canton of Fribourg and the Swiss National Science Foundation (to CDV), and grants from the National Natural Science Foundation of China (31370015 and 31230017) and the Chinese Academy of Sciences (XDB08010302 and Youth Innovation Promotion Association CAS) to JD and TZ.
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
TAP pull-down experiments showing that the reduced interaction between Gtr1-TAP and Gtr2-V5 in an ego2Δ strain can be rescued by re-introduction of EGO2 expressed from a plasmid. (PDF 102 kb)
Supplementary information, Figure S2
In vitro functional analyses of the Ego1-Ego2-Ego3 ternary complex (EGO-TC). (PDF 183 kb)
Supplementary information, Figure S3
Structural comparisons of the Ego proteins and several representative Roadblock domain-containing proteins. (PDF 103 kb)
Supplementary information, Table S1
Strains used in this study. (PDF 261 kb)
Supplementary information, Table S2
Plasmids used in this study. (PDF 184 kb)
Rights and permissions
About this article
Cite this article
Powis, K., Zhang, T., Panchaud, N. et al. Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling. Cell Res 25, 1043–1059 (2015). https://doi.org/10.1038/cr.2015.86
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2015.86
Keywords
This article is cited by
-
A nutrient-induced affinity switch controls mTORC1 activation by its Rag GTPase–Ragulator lysosomal scaffold
Nature Cell Biology (2018)
-
Signal integration in the (m)TORC1 growth pathway
Frontiers in Biology (2018)
-
Structural insight into the Ragulator complex which anchors mTORC1 to the lysosomal membrane
Cell Discovery (2017)
-
Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1
Nature Communications (2017)
-
Structural basis for the assembly of the Ragulator-Rag GTPase complex
Nature Communications (2017)