Abstract
Interactions between the BCL-2 family proteins determine the cell's fate to live or die. How they interact with each other to regulate apoptosis remains as an unsettled central issue. So far, the antiapoptotic BCL-2 proteins are thought to interact with BAX weakly, but the physiological significance of this interaction has been vague. Herein, we show that recombinant BCL-2 and BCL-w interact potently with a BCL-2 homology (BH) 3 domain-containing peptide derived from BAX, exhibiting the dissociation constants of 15 and 23 nM, respectively. To clarify the basis for this strong interaction, we determined the three-dimensional structure of a complex of BCL-2 with a BAX peptide spanning its BH3 domain. It revealed that their interactions extended beyond the canonical BH3 domain and involved three nonconserved charged residues of BAX. A novel BAX variant, containing the alanine substitution of these three residues, had greatly impaired affinity for BCL-2 and BCL-w, but was otherwise indistinguishable from wild-type BAX. Critically, the apoptotic activity of the BAX variant could not be restrained by BCL-2 and BCL-w, pointing that the observed tight interactions are critical for regulating BAX activation. We also comprehensively quantified the binding affinities between the three BCL-2 subfamily proteins. Collectively, the data show that due to the high affinity of BAX for BCL-2, BCL-w and A1, and of BAK for BCL-XL, MCL-1 and A1, only a subset of BH3-only proteins, commonly including BIM, BID and PUMA, could be expected to free BAX or BAK from the antiapoptotic BCL-2 proteins to elicit apoptosis.
Similar content being viewed by others
Introduction
The BCL-2 family proteins are central regulators of the mitochondrion-mediated apoptotic cell death. They are characterized by containing up to four conserved stretches of amino acids, known as BCL-2 homology (BH) domains 1, 2. They are usually grouped into three distinct subclasses. One subclass is composed of BAX and BAK that mediate apoptosis by triggering destabilization of the outer mitochondrial membrane (OMM) and consequently releasing the apoptogenic factors, such as cytochrome c, from mitochondria to the cytosol 3, 4. Another subclass is composed of the BH3-only proteins (including BIM, BAD, PUMA and Noxa) that sense and convey pro-death signals and ultimately activate downstream BAX and BAK 5, 6. While BAX and BAK contain the BH1 through BH3 domains and are homologous to each other, the BH3-only proteins are unrelated to each other except that they all contain the BH3 domain. The activation of BAX/BAK is suppressed by the remaining subclass composed of BCL-2, BCL-XL, BCL-w, MCL-1, A1 and BCL-B, all of which contain the four BH domains. The three-dimensional structures of BCL-XL in complex with a BH3 domain-containing segment derived from BAK, BIM or BAD revealed that each fragment binds to an extended hydrophobic groove in BCL-XL, known as the BH3-binding groove 7, 8, 9.
The interplay between the three subclasses of the BCL-2 family determines the fate of cells in response to developmental or stress signals. The mechanism of how BAX and BAK are activated by the BH3-only proteins in dying cells has been of intense investigation. Two main models have been put forth. A widely accepted model is the direct activation model, which suggests that the “sensitizer” or “inactivator” BH3-only proteins (BAD, BIK, BMF, Hrk and Noxa) release the “activator” BH3-only proteins (BIM, BID and possibly PUMA) sequestered by the antiapoptotic BCL-2 subfamily members, and that these “activators” are required for activating inert BAX/BAK via a direct but transient binding interaction 5, 10, 11, 12, 13. The other model, called indirect activation model, suggests that the antiapoptotic BCL-2 proteins inhibit apoptosis by sequestering a small proportion of the activated BAX and BAK in healthy cells, and that a subset of the BH3-only proteins, including the “activators”, engages the antiapoptotic proteins to release BAX and BAK in dying cells 6, 14, 15, 16. In this model, the “activator” BH3-only proteins are not directly involved in the activation of BAX/BAK. The indirect model is opposed in the field mainly by two reasons. First, BAX is mostly cytosolic and monomeric, with a minor fraction bound to the OMM 17, where most of the antiapoptotic BCL-2 proteins are found in healthy cells. Second, BAX has been considered to interact with the antiapoptotic BCL-2 proteins only weakly, if it does, and therefore the significance of these interactions has been elusive 16, 18, 19. Nevertheless, mutational and other studies strongly support the view that the antiapoptotic proteins should be able to engage the BH3 domain of BAX to prevent it from mediating apoptosis 6, 16.
By isothermal titration calorimetry (ITC), we found that BCL-2 and BCL-w bind to a 36-mer BH3 peptide of BAX with potent affinity. Subsequently, we determined the structure of BCL-2 in complex with a BAX peptide, which represents the first structure of BCL-2 bound to a BH3 peptide and also the first structure of the BAX peptide in complex with a BCL-2 family protein. The structure enabled us to rationally design a full-length BAX mutant with significantly reduced affinity for BCL-2 by introducing triple point mutations within the BAX BH3 domain. Phenotypic characterization of this BAX variant by various cell-based assays indicates that BCL-2 and BCL-w inhibit BAX-mediated apoptosis by tightly binding to BAX. Finally, by comprehensive quantification of the binding interactions between the antiapoptotic BCL-2 proteins and BH3 peptides derived from the BH3-only proteins, we show that a subset of the BH3-only proteins bind even more tightly to BCL-2 and BCL-w, suggesting that they should be able to liberate BAX from the sequestration by the two antiapoptotic proteins. Likewise, the delineated hierarchical binding potency points the identity of BH3-only proteins that can disrupt the interactions between other BCL-2 proteins and BAX or BAK.
Results
Preferential binding affinity of BCL-2 and BCL-w for BAX over BAK
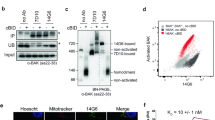
In order to investigate the interactions between the antiapoptotic BCL-2 subfamily and the apoptosis mediators BAX and BAK, we produced five different recombinant mouse or human antiapoptotic BCL-2 relatives (BCL-2, BCL-XL, BCL-w, MCL-1 and A1) and quantified their binding affinity for a 36-mer peptide containing the BH3 domain of BAX or BAK by ITC that yields apparent dissociation constants (KD). The use of the 36-mer BH3 peptides was based on the ample structural, biochemical and cellular data demonstrating that a BH3 domain-containing segment with < 34 amino acids of the BH3-only proteins is responsible for their interactions with the antiapoptotic BCL-2 relatives 8, 9, 15, 20, 21. The quantification showed that BCL-2 and BCL-w interact tightly with the BAX peptide, exhibiting the KD of 15.1 and 22.9 nM, respectively, while BCL-XL and MCL-1 bind the peptide with lower affinity (Figure 1A). The potent affinity between BCL-2 and the BAX peptide is in contrast with the reported data in the literature showing that a 21-mer or 26-mer BAX peptide interacts with BCL-2, but only very weakly 18, 19. In contrast to the BAX peptide, the BAK peptide interacted with BCL-XL and MCL-1 preferentially (8.1 and 1.3 nM, respectively) over BCL-2 and BCL-w (Figure 1A). Our quantification is consistent with the reported observation that endogenous BAK was immunoprecipitated with transiently expressed full-length BCL-XL and MCL-1, but not with full-length BCL-2 and BCL-w 22. Herein, we further show that endogenous BAX associates mainly with endogenous BCL-2 and also with BCL-w in human embryonic kidney 293T cells, and confirm that endogenous BAK associates predominantly with endogenous BCL-XL and MCL-1 (Figure 1B). However, our quantification is quite inconsistent with a previous observation that BCL-2 and BCL-XL interact with a 34-mer BAX peptide with similar affinities, IC50 of 0.10 and 0.13 μM, respectively 16. For confirmation, a native gel-based qualitative binding assay was carried out, which showed that the 36-mer BAX peptide interacts with BCL-2 more tightly than it does with BCL-XL (Figure 1C). Our data conclusively show that BCL-2 and BCL-w bind BAX preferentially over BAK. For in vitro binding assays, we used the detergent Triton X-100 (0.5%), which is known to alter the conformation of BAX and BAK and is therefore widely employed to detect the interaction between the two apoptotic mediators and the antiapoptotic BCL-2 relatives 15, 16, 22, 23.
Interactions between antiapoptotic BCL-2 proteins and BAX/BAK. (A) Binding affinities. ITC analysis was carried out by titrating the 36-mer BH3 peptides of BAX and BAK (0.2 mM) into the indicated antiapoptotic BCL-2 proteins (10 μM). The KD values were deduced from curve fittings of the integrated heat per mol of added ligand and are listed in the tables. Two representative ITC runs are shown below. (B) Endogenous interactions. Whole cell lysates of 293T cells were used for immunoprecipitation with control rabbit serum, anti-BAX or anti-BAK antibody followed by immunoblotting with antibodies against the indicated BCL-2 proteins. (C) The BAX peptide preferentially binds to BCL-2 over BCL-XL. The BAX peptide (5 μM) was incubated together with BCL-2 (lane 2), BCL-XL (lane 4) or both the proteins (lane 6) at the same concentration (5 μM), and the mixture was visualized on a native gel. Free BCL-2 is unobservable, while most of BCL-XL remains free. (D) A1 binds both endogenous BAX and BAK. HA-tagged A1 was transiently expressed in 293T cells and whole-cell lysates were used for immunoprecipitation with control rabbit serum, anti-BAX or anti-BAK followed by immunoblotting with anti-HA, anti-BAX or anti-BAK antibody.
A1 binds both BAX and BAK potently
The interplay between A1 and BAX/BAK has been controversial to date 24, 25, 26. Recently, transiently expressed A1 was reported to interact with overexpressed BAX and endogenous BAK, but not with endogenous BAX in HeLa cells or immortal baby mouse kidney cells 24. We show here that A1 interacts with both the BAX and BAK peptides potently (KD of 3-4 nM; Figure 1A). We also performed cell-based assay to detect an interaction between full-length versions of the proteins. Transiently expressed A1 bound both endogenous BAX and BAK in 293T cells (Figure 1D), which is consistent with the measured potent binding affinity. In contrast, we were unable to detect an interaction between transiently expressed BAX or BAK with endogenous A1 in 293T cells. However, it is possible that the endogenous expression of A1 is simply too low to be detected in these cells, as was the case in mouse embryonic fibroblast (MEF) cells 22.
Structure of BCL-2 in complex with a 31-mer BAX peptide
Inspired by our quantification, we subsequently determined the structure of BCL-2 bound to a 31-mer BAX peptide at 2.7 Å resolution (Supplementary information, Table S1). The BAX peptide forms an amphipathic α-helix and binds to the BH3-binding groove of BCL-2, as commonly observed in all the other reported structures of antiapoptotic BCL-2 proteins in complex with a BH3 peptide (Figure 2 and Supplementary information, Figure S1A). Although N-terminal five residues of the BAX peptide are invisible in the structure, the visible portion alone spans the entire BH3-binding groove, and is much longer than the conventionally defined BH3 domain (Figure 2). We note that the 31-mer BAX peptide (residues 52-82) binds BCL-2 nearly as potently as the 36-mer BAX peptide, but a 26-mer BAX peptide (residues 52-77) has poor binding affinity (KD of 610 nM) for BCL-2 (Figure 2). The difference in the binding affinity appears to arise from direct intermolecular interactions and increased helical propensity involving the last five residues (Arg78-Asp82) present in the 31-mer, but not in the 26-mer peptide. Arg78 is involved in the hydrophilic interaction with BCL-2 and Ala81 in the van der Waals interaction (interatomic distances < 3.8 Å). While the rest three residues do not interact with BCL-2 directly, they are the last part of the BAX α-helix at the binding groove, indicating that they contribute to the binding affinity by increasing the helical propensity of the BAX peptide. Consistently, circular dichroism spectroscopic analysis showed that the helical content of the 31-mer peptide (46%) is higher than that of the 26-mer peptide (37%) in 30% trifluoroethanol. Our observation and interpretation explain, at least partly, the low-affinity interactions between BAX peptides and BCL-2 found in the literature. The BCL-2-BAX peptide complex involves hydrophobic interactions mediated by five nonpolar BAX residues, four of which are highly conserved in the proapoptotic BCL-2 proteins (Figure 3A, left panel; Figure 3B). Alanine substitution of the nonconserved residue, Met74, reduced the affinity of the BAX peptide for BCL-2 from 15 nM to 203 nM in KD values, which confirms its contribution to the binding interaction. Notably, the interaction between BCL-2 and the BAX peptide involves five intermolecular salt bridges (Figure 3A, right panel). This is a distinctive feature in comparison with the 10 other available structures of the antiapoptotic BCL-2 proteins in complex with a BH3 peptide, which contain no more than three intermolecular ionic interactions (Supplementary information, Figure S1B). The five BAX residues engaged in the salt bridges are Glu61, Arg64, Asp68, Glu69 and Arg78. Of these, Asp68 and Glu69 are conserved in the BH3 domains, but the other three charged residues are not (Figure 3B). These nonconserved residues pair with Arg139, Asp140 or Glu200 of BCL-2. The importance of these ionic interactions was estimated by individually substituting the five charged residues of BAX with alanine and measuring the binding affinity of the resulting mutated peptides for BCL-2. Each of the mutations reduced the binding affinity in varying degrees, with the maximum reduction by the D68A mutation (Figure 3C). Notably, triple mutations of the nonconserved residues (E61A, R64A and R78A) caused ∼52-fold reduction in the binding affinity (Figure 3C), demonstrating that they together contribute greatly to the binding affinity of BAX for BCL-2. The triple mutations also drastically reduced the affinities of the BAX peptide for BCL-w from 23 nM to 943 nM and for BCL-XL from 255 nM to 3.57 μM in KD values, indicating that these charged residues are commonly important for interacting with the antiapoptotic BCL-2 proteins.
Interactions between BCL-2 and BAX BH3 peptides. (Left) The structure of BCL-2 bound to the 31-mer BAX peptide. BCL-2 and the BAX peptide are in pink and green, respectively. (Right) Binding affinities. Each of the three indicated BAX peptides was titrated into the BCL-2 solution and the deduced KD values are shown in the table. The sequences of the BAX peptides are listed below, with the conventionally defined BH3 domain marked in red.
Structural and mutational analysis of the interaction between BCL-2 and the BH3 domain of BAX. (A) Intermolecular interactions. Two surface models of BCL-2 with the bound BAX peptide in green, showing the hydrophobic (left) and the ionic interactions (right) separately. The residues engaged in those interactions are shown in sticks and labeled. The surface coloring scheme is as follows: yellow for Val, Leu, Ile, Tyr, Phe, Trp, Met and Ala; blue for Lys, Arg and His; red for Glu and Asp; gray for other amino acids. (B) Sequence alignment of the BH3 domains in the proapoptotic BCL-2 proteins. Conserved residues are highlighted in red (> 80% similarity) or pink (> 60% similarity) columns. The black and red arrows indicate the BAX residues shown in (A). The five consensus BH3 residues that are known to be critical for the interactions with the antiapoptotic BCL-2 family members 7 are indicated by asterisks. (C) Contribution of the charged residues of BAX to the binding affinity. BAX peptides (36-mer) with the alanine substitution of the indicated charged residues were titrated into BCL-2, and the deduced KD values are listed in the table. Shown in the bottom is the ITC run for the titration of the BAX peptide with the triple substitutions.
Generation of an inducibly active BAX mutant with impaired affinity for BCL-2
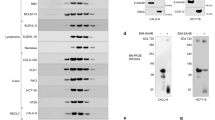
With the aim to show that the biochemically and crystallographically defined interaction between recombinant BCL-2 and the BAX peptide is physiologically relevant, we sought to generate a full-length BAX variant that has impaired binding affinity for full-length, wild-type BCL-2, but retains all the other properties of wild-type BAX. A comparison between the presented structure and the structure of free BAX 27 shows that the BAX peptide bound to BCL-2 corresponds to α2 and α3 of BAX and that the nonconserved, charged BCL-2-interacting residues of BAX (Glu61, Lys64 and Arg78) are all solvent-exposed in free BAX (Supplementary information, Figure S2). Therefore, substitution of these residues with alanine was expected to impair the affinity for BCL-2 without affecting the stability of the soluble BAX conformer. We generated a full-length BAX variant containing the same triple mutations, which is referred to as BAX(AAA), and performed cell-based protein-binding assays. Whereas full-length wild-type BAX clearly interacted with endogenous BCL-2 in 293T cells, the BAX variant failed to show a detectable interaction, demonstrating that the three charged residues are important for BCL-2 binding in cells (Figure 4A). We also generated a full-length BAX mutant containing a substitution of Leu63 with alanine on the BH3 domain. Leu63, an invariant residue in many BH3 domains (Figure 3B), is involved in tight intermolecular hydrophobic interactions in the structure of the BCL-2-BAX peptide complex (Figure 3A). As expected, the BAX(L63A) mutant did not detectably interact with endogenous BCL-2 in cells (Figure 4A), like BAX(AAA). BAX(L63A) was used as a negative control for its inactivity, as described below.
BAX(AAA) has impaired affinity for BCL-2 and is functionally active. (A) Cell-based protein-binding assay. The indicated proteins were transiently expressed in 293T cells. The amount of BCL-2 or endogenous BAX associated with these proteins was detected by immunoprecipitation using antibody against Flag followed by immunoblot analysis using antibody against BCL-2 or BAX. BAX(AAA) fails to bind endogenous BCL-2, but associates with endogenous BAX. Coprecipitation of Flag-BAX and endogenous BAX is conceivably through homooligomerization. Transfection with empty vector as a control is indicated by “Vector” throughout the figures. (B, C) Apoptosis assays. Bax−/− Bak−/− MEFs stably expressing the indicated proteins were treated with etoposide (100 μM) for 36 h, and cell viability or the ratio of apoptotic cells was assessed by PI staining (B) or Annexin V staining (C) and flow cytometry. Values are the average of three independent assays ± standard deviations. The two assays clearly show that BAX(AAA) induces apoptosis as efficiently as wild-type BAX. Further corroboration of these results is shown in Supplementary information, Figure S3. Expression levels of the BAX proteins are shown in Supplementary information, Figure S4A.
Importantly, BAX(AAA) is fully proapoptotic and remains inducibly active, like wild-type BAX and the previously described mutants BAX K64A 6 and BAX D68R 16. This conclusion is supported by several types of experiment. First, like wild-type BAX, BAX(AAA) could associate with endogenous BAX (Figure 4A). Second, BAX(AAA) did not induce substantial spontaneous cell death when stably overexpressed in Bax, Bak double-deficient (Bax−/− Bak−/−) MEFs, but was as potent as wild-type BAX when those cells received different apoptotic stimuli (Figures 4B, 4C and 5 and Supplementary information, Figure S3A). Finally, when those cells received an apoptotic stimulus but not before, BAX(AAA), like wild-type BAX, exhibited a conformational change detectable by the conformation-specific antibody 6A7 (Supplementary information, Figure S3B), which binds to the N-terminus of BAX exposed upon BAX activation 28. In these experiments, the control mutant BAX(L63A) was not reactive to 6A7 (Supplementary information, Figure S3B), did not detectably oligomerize with endogenous BAX (Figure 4A) and exhibited greatly impaired apoptotic activity (Figures 4B, 4C and 5 and Supplementary information, Figure S3A), showing that BAX(L63A) is functionally defective. It is noted that the amount of immnoprecipitated Flag-tagged BAX(L63A) was far less abundant than Flag-tagged BAX or BAX(AAA) (Figure 4A, second panel), although the expression level of the three proteins was similar (Figure 4A, third panel). We posit that this is because BAX(L63A) cannot form an oligomer, while wild-type BAX and BAX(AAA) can oligomerize and be more efficiently precipitated by immunoprecipitation than a monomeric form of the protein. Probably, Leu63 is a critical residue for BAX to homooligomerize to form a protein-conducting channel on the OMM. In other cell-based experiments with BAX(AAA), we employed this mutant as a negative control that cannot induce apoptosis.
BAX(AAA)-mediated apoptosis is barely inhibited by BCL-2 and BCL-w. (A) Bax−/− Bak−/− MEFs stably expressing HA-tagged BCL-2 (DKO.BCL-2) were transfected with a BAX- or BAX(AAA)-expression vector and treated with 50 μM etoposide for 36 h. The apoptotic cells were estimated by PI staining and flow cytometry. (B) At 16 h post-transfection with a plasmid expressing HA-BCL-2, the Bax−/− Bak−/− MEF cells stably expressing vector only, BAX, BAX(AAA) or BAX(L63A) were treated with 100 μM etoposide for 8 h, fixed, and were immunostained with anti-HA antibody (red) or 6A7 (green) followed by confocal analysis. A single representative optical section is presented. The white and yellow arrows indicate BCL-2-expressing cells and 6A7-positive cells (Act BAX), respectively. The bottom panel shows the percentage of BCL-2-transfected cells positively stained with 6A7 antibody (∼200 cells collected per sample). (C) Bax−/−Bak−/− MEFs stably expressing vector only, BAX, BAX(AAA) or BAX(L63A) were transfected for ectopic expression of HA-tagged BCL-w. After etoposide (100 μM) treatment for 36 h, cells were stained with Annexin V and analyzed by flow cytometry (top). Apoptotic cells are estimated and plotted (bottom). The error bars indicate standard deviations calculated from three independent assays. Expression levels of the BAX proteins and BCL-w are shown in Supplementary information, Figure S4C.
BAX(AAA) is refractory to inhibition by BCL-2 and BCL-w
We next examined whether the apoptotic activity of BAX or BAX(AAA) can be restrained by BCL-2 or BCL-w. Coexpression of BCL-2 with them in Bax−/− Bak−/− MEFs significantly attenuated the BAX-mediated apoptosis upon etoposide treatment, but it had minimal effect on the BAX(AAA)-mediated apoptosis (Figure 5A). This difference was not due to an enhanced level of BAX(AAA) expression, as equivalent amounts of BAX and BAX(AAA) were detected in the stable cell lines (Supplementary information, Figure S4B). To corroborate this result, we examined the individual cells treated with the 6A7 antibody. Confocal immunofluorescence microscopy showed that 6A7-positive BAX coexisted with BCL-2 in BAX(AAA)-expressing cells, but not in BAX- or BAX(L63A)-expressing cells (Figure 5B), indicating that BCL-2 cannot block BAX(AAA) activation whereas it can block the activation of wild-type BAX. A counting showed that over 60% of the BCL-2-transfected BAX(AAA)-expressing cells were positive to the 6A7 antibody (Figure 5B). In comparison, the BCL-2-transfected BAX-expressing cells rarely exhibited the activated BAX, and these few 6A7-positive cells seldom contained BCL-2 (Figure 5B). In parallel, we also examined whether BCL-w is able to inhibit the apoptotic activity of BAX or BAX(AAA) in Bax−/− Bak−/− MEFs. Ectopic expression of BCL-w protected the BAX-expressing cells but not the BAX(AAA)-expressing cells from undergoing apoptosis (Figure 5C), as observed with BCL-2. Together, these cell-based phenotypic analyses of BAX(AAA) demonstrate that the crystallographically defined interaction between BCL-2 and the BAX peptide is physiologically relevant, and that the tight binding of BCL-2 or BCL-w to BAX is critical for inhibiting BAX-mediated apoptosis.
Binding affinities between antiapoptotic BCL-2 relatives and BH3-only proteins
In order to know whether the observed interactions between the antiapoptotic BCL-2 proteins and BAX/BAK can be disrupted by certain BH3-only proteins, we determined the binding affinities for the interactions between the five antiapoptotic BCL-2 relatives and 36-mer BH3 peptides derived from eight well-studied BH3-only proteins (BIM, BID, PUMA, BAD, BIK, BMF, Hrk and Noxa). The BIM, BID and PUMA peptides interacted extremely tightly with all of the five antiapoptotic BCL-2 proteins (KD < 8.9 nM; Figure 6A). These binding affinities are higher than that determined for the rest of the peptides derived from BAD, BIK, BMF, Hrk and Noxa (KD ≥ 10.9 nM), except for the BIK peptide exhibiting comparable affinity for BCL-w (KD of 4.4 nM; Figure 6A). This data set soundly supports the scheme of classifying the BH3-only proteins into two subgroups: “promiscuous” ones (BIM, BID and PUMA) and “selective” ones (BAD, BIK, BMF, Hrk and Noxa) based on the measurement of binding affinity on a relative scale 20. The binding affinities determined in this study are on the absolute scale, and therefore can be directly compared with each other. Accordingly, the combination of the two data sets (Figures 1A and 6A) readily shows the hierarchy of the quantified binding affinities (Figure 6B). The established hierarchy readily indicates which BH3-only protein would be able to free BAX or BAK, if they are sequestered by an antiapoptotic BCL-2 protein. For example, BIM, truncated BID (tBID) and PUMA should be able to readily displace BAX or BAK from any antiapoptotic BCL-2 proteins, while BAD could displace BAX from BCL-2 but less efficiently compared with the three BH3-only proteins. Our quantification is inconsistent with the reported observation that a 34-mer BAX peptide interacts with BCL-2 much more weakly than a BAD or a BMF BH3 peptide does 16, 20.
Interactions between antiapoptotic BCL-2 proteins and BH3-only proteins. (A) Binding affinities. ITC analysis was performed by titrating the 36-mer BH3 peptides derived from the indicated BH3-only proteins (0.2 mM) into the five antiapoptotic BCL-2 proteins (10 μM). (B) A plot of the measured KD values on a log scale. The diagram includes the KD values measured for the BAX and BAK peptides (Figure 1A). The dash marks in front of the names of the proapoptotic proteins (in black or red letters) are positioned at the measured binding affinities of their BH3 peptides for the indicated antiapoptotic BCL-2 proteins (in blue letters).
To test the validity of the hierarchy of the binding affinities, we performed a competition assay in which a complex between an antiapoptotic BCL-2 protein and a BH3 peptide was challenged by another BH3 peptide. The BAD peptide was hardly able to displace the BIM peptide from BCL-2 even at eight-fold molar excess (Figure 7A), which is consistent with the positions of BIM and BAD in the hierarchy. Likewise, the BIM peptide easily displaced the BAX peptide bound to BCL-2 even at 1:8 molar ratio between the BIM and the BAX peptides (Figure 7B and 7C), while the BAD peptide at eight-fold molar excess over the BAX peptide could not displace completely the BAX peptide from BCL-2 (Figure 7B and 7D). We also examined whether transiently expressed Flag-tagged BIMEL (the most abundant BIM isoform), BAD or Noxa, might disrupt the endogenous BCL-2-BAX interaction in 293T cells. BIMEL and BAD, but not Noxa, dissociated the BCL-2-BAX complex and formed a complex with BCL-2 (Figure 7E), which is consistent with the hierarchy of the binding affinities.
Displacement assays. For in vitro assay (A-D), the 36-mer BH3 peptides were used, which are indicated by the names of the proapoptotic BCL-2 proteins. BCL-2 was preincubated with a BH3 peptide at 1:1 molar ratio or up to 1:8 molar ratio (in C) for 1 h. A competitor peptide (in red) was added to the mixture and incubated for additional 1 h. The numbers in parentheses indicate the ratios between the peptides. The mixture was visualized on a native gel. (A) The BAD peptide hardly displaced the BIM peptide bound to BCL-2, as most of the BCL-2-BIM peptide complex remained intact up to eight-fold molar excess of the BAD peptide over the BIM peptide. (B) The BIM peptide displaced the BAX peptide completely and formed a complex with BCL-2 (lane 6). The BAD peptide displaced the BAX peptide but only slightly (lane 7). The NoxaA peptide failed to displace the BAX peptide (lane 8). (C) The BIM peptide readily displaced the BAX peptide bound to BCL-2. Formation of BCL-2-BIM peptide complex is visible even at 8:1 molar ratio between the BAX and BIM peptides. (D) The BAD peptide inefficiently displaced the BAX peptide bound to BCL-2. The BCL-2-BAX peptide complex remained even at 1:8 molar ratio between the BAX and BAD peptides. (E) Cell-based displacement assay. Flag-BIMEL, Flag-BAD or Flag-Noxa was transiently expressed in 293T cells and the effect of the expression on the endogenous BCL-2-BAX interaction was assessed by immunoprecipitation and immunoblotting. (Left) BIMEL sequesters BCL-2 from BAX binding; (middle) BAD binds BCL-2 and displaces BAX; (right) Noxa does not bind BCL-2 and has no effect on the BCL-2-BAX interaction. The asterisks indicate the light chain of antibody.
Discussion
Preferential interactions between antiapoptotic BCL-2 proteins and BAX or BAK
While BAX was originally identified as a BCL-2-binding protein 29, 30, its interaction has been believed to be weak. Our binding affinity measurement shows that the BAX BH3 peptide indeed binds to BCL-2 and BCL-w potently, and that it also binds to BCL-XL and MCL-1, but with lower affinity. This preferential binding affinity is opposite to that observed with the BAK peptide, which binds to BCL-XL and MCL-1 with at least nine times higher affinity than it binds to BCL-2 and BCL-w. The preferential intermolecular interactions plausibly explain a number of previously reported observations in the context of the indirect activation model. For example, overexpression of MCL-1 was reported to block staurosporine-induced apoptotic cell death in Bax−/− but not in Bak−/− immortal baby mouse kidney cells 24. These observations are readily understandable, because both BCL-XL and MCL-1 have potent binding affinity for BAK, while they bind BAX with ∼30-fold lower affinity (Figure 1A). For another example, the BH3 mimetic ABT-737, which has high affinity for BCL-2, BCL-XL and BCL-w, but not for MCL-1, cannot induce BAX-mediated apoptosis in wild-type or Bak−/− MEF cells unless MCL-1 is inhibited by Noxa overexpression 6. This observation, indicating that MCL-1 is also able to neutralize BAX, appears contradictory to the finding that MCL-1 does not bind endogenous BAX. However, the two observations can be reconciled by the fairly high, albeit not potent, binding affinity between MCL-1 and the BAX peptide (KD of 40 nM), which should enable MCL-1 to sequester BAX when the other BCL-2 relatives are neutralized.
A membrane-bound form of BAX would be the target for the BCL-2 relatives
The most important observation made in this study is that the delineated interactions between BCL-2 and BCL-w with BAX are physiologically relevant. How and where would these interactions take place in cells? While the BH3 domain-containing segment of BAX is responsible for binding to BCL-2, this region is at least half-buried inside the structure of isolated BAX (Supplementary information, Figure S2). Likewise, the BAK BH3 peptide used in this study is also half-buried inside the protein in the structure of isolated BAK 31. Therefore, the antiapoptotic BCL-2 proteins should be unable to bind BAX or BAK, unless their BH3 domain is exposed as a result of drastic conformational change. It is known that a minor fraction of BAX exists on the OMM 17. On the membrane, BAX is believed to proceed through multiple conformational states, until they form homooligomerized protein-conducting channels 32. Conceivably, one of the conformational states of BAX exposes its BH3 domain and is accessible for binding by the BCL-2 relatives. Unlike BAX, most or at least some BAK is pre-bound to BCL-XL and MCL-1 on the OMM 22, 33, and the binding is dependent on the BH3 domain of BAK 7, 22, 34. The conformational state of BAK that is competent for binding to the antiapoptotic proteins is dubbed as “primed form” 6, 14. These and our data suggest that a membrane-bound form of both BAX and BAK is the target for binding by antiapoptotic BCL-2 proteins, and that BAX is sequestered mainly by BCL-2 and BCL-w, while BAK is sequestered mainly by BCL-XL and MCL-1.
Implications for BAX activation in apoptotic cells
The indirect activation model argues that some of the BH3-only proteins should be able to bind BCL-2, BCL-w or A1 more potently than BAX does in order to displace BAX from these antiapoptotic proteins in cells undergoing apoptosis. Indeed, our quantification and cell-based assays indicate that a subset of the BH3-only proteins, commonly including BIM, tBID and PUMA, are able to readily displace BAX bound to any of the three antiapoptotic proteins (Figure 6B). The liberated BAX may then be able to undergo activational conformational change. Activated BAX was shown to recruit cytosolic BAX to membranes and activate it 35. This autonomous BAX activation appears to be supported by the constitutively active BAX mutants reported in the literature, which contain L70A or D71A mutation and exhibit a reversed distribution of BAX in cells 36. The two residues are buried in free BAX, and therefore these mutations presumably affect the stability of the soluble BAX conformer, promoting translocation of the resulting BAX mutants to the OMM where they could be autoactivated. Critically, BAX can be normally activated in Bim−/−Bid−/− MEFs 6, suggesting that the “activator” BH3-only proteins are dispensable for BAX activation. On the contrary, numerous studies have indicated the involvement of BIM or tBID in BAX activation; they are believed to act by interacting with BAX transiently 37, 38 and by promoting membrane insertion of BAX 39. A reconciling senario would be that BIM or tBID is mobilized under apoptotic stimuli at exceedingly high concentration compared with that of neutralizing BCL-2 proteins and then they not only liberate BAX from sequestration by the antiapoptotic BCL-2 proteins but also accelerate BAX activation via direct interaction. Less amount of BIM or tBID should be necessary, if other BH3-only proteins, such as BAD, are simultaneously mobilized.
Conclusions
In this work, we biochemically and structurally characterized previously unrecognized tight interaction of BAX with BCL-2 and BCL-w, which are shown to be critical for blocking BAX activation. We expect that the hierarchical binding potency reported herein will serve as decisive information for interpreting various data in this field. Finally, the interaction between BCL-2 and the BAX peptide delineated herein forms a foundation for designing cancer therapeutic agents that selectively inhibit BCL-2 or BCL-w.
Materials and Methods
Purification of BCL-2 family proteins
Each of the DNA fragments coding for mouse BCL-XL (residues 1-44 and 85-196), BCL-w (residues 6-152) and MCL-1 (residues 152-308) was cloned into pProEX HTa (Invitrogen). A chimeric human BCL-2 (residues 1-50 and 92-207) was constructed by removing the internal long loop (residues 51-91) and replacing residues 35-50 with residues 33-48 of BCL-XL to solubilize the protein as reported earlier 40. The DNA fragment coding for mouse A1 (residues 1-152) was cloned into pGEX-4T-3 (GE Healthcare). The protein contains two mutated residues (P104K and C113S) that improve the solubility of the protein 41. The proteins were expressed in the E. coli BL21(DE3) strain (Novagen) at 18 °C overnight and were purified using a Ni-NTA column (QIAGEN) or a glutathione-agarose column (Peptron) and a HiTrap Q anion exchange column (GE Healthcare). The N-terminal glutathione S-transferase tag fused to A1 was cleaved with TEV protease after the glutathione-agarose column purification step and was removed from A1 by a HiTrap Q anion exchange column.
Peptides
Synthetic 36-mer peptides derived from mouse BIM (residues 136-171), BID (residues 76-111), PUMA (residues 127-162), BAD (residues 137-172), BIK (residues 41-76), BMF (residues 210-245), Hrk (residues 23-58), NoxaA (residues 13-48), NoxaB (residues 65-100), BAK (residues 62-97) and BAX (residues 49-84) were purchased from Peptron (Korea), Anygen (Korea) or GenScript (USA). Also purchased were BAX peptides of 26-mer (residues 52-77) and 31-mer (residues 52-82) and mutated BAX peptides of 36-mer (residues 49-84) containing a single E61A, R64A, D68A, E69A or R78A mutation or triple E61A/R64A/R78A mutations. The helical content of the BAX peptides was estimated by circular dichroism as previously described 21.
Isothermal titration calorimetry
All measurements were carried out at 25 °C on a MicroCalorimetry System (MicroCal). Protein samples were dialyzed against the solution containing 20 mM Tris-HCl (pH 7.5) and 50 mM NaCl. The samples were degassed for 20 min and centrifuged to remove any residuals prior to the measurements. Dilution enthalpies were measured in separate experiments (titrant into buffer) and subtracted from the enthalpies of the binding between the protein and the titrant. Data were analyzed using the Origin software (OriginLab Corp.).
Preparation and structure determination of the BCL-2-BAX peptide complex
The chimeric BCL-2, referred to as BCL-2, was expressed in the E. coli BL21(DE3) RIG strain (Novagen) at 18 °C overnight and purified using a Ni-NTA column, a HiTrap Q anion exchange column and a HiLoad 26/60 Superdex 75 gel filtration column (GE Healthcare), equilibrated with 20 mM Tris-HCl (pH 7.5), 50 mM NaCl and 1 mM dithiothreitol. The final BCL-2 sample was mixed with the 31-mer BAX peptide (residues 52-82), and the crystals of the resulting complex were obtained by the hanging-drop vapor diffusion method at 24 °C by mixing and equilibrating 1 μl of each of the protein solution (10 mg/ml) and a precipitant solution containing 0.1 M sodium acetate (pH 4.2) and 3.0 M sodium formate. Before data collection, the crystals were immersed briefly in a cryoprotectant solution, which was the reservoir solution, plus 5% glycerol. A diffraction data set at 2.7 Å was collected on the beam line 4A at the Pohang Accelerator Laboratory, Korea. The structure was determined by the molecular replacement method with the CCP4 version of MolRep 42 using the structure of BCL-2 40 as a search model. Subsequently, model building and refinement were carried out using the programs COOT 43 and CNS 44. The final model does not include residues 1-9, 32-50 (including the entire BCL-XL substitution region) and 207 of BCL-2 and residues 52-56 of BAX, whose electron densities were not observed or were very weak. Crystallographic data statistics are summarized in Supplementary information, Table S1.
Cell culture and retroviral infection
Bax−/− Bak−/− MEFs and 293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine and 1% penicillin-streptomycin (Gibco-BRL). Transient transfection was performed with Fugene 6 (Roche), Lipofectamine 2000 (Invitrogen) or calcium phosphate (Clontech). Bax−/− Bak−/− MEF stable cell lines were established by retroviral infection. pBabe/puro retroviral constructs encoding BAX proteins were transiently transfected into HEK293 EcoPack packaging cells (BD Biosciences) and viral supernatants were used to infect cells followed by the selection with 2 μg/ml of puromycin (Sigma-Aldrich).
Immunoblotting and immunoprecipitation
For immunoblotting, the polypeptides were resolved by SDS-PAGE and transferred onto a PVDF membrane (Bio-Rad). The membranes were blocked with 5% non-fat milk and probed with antibodies. Goat antibodies coupled to horseradish peroxidase specific to mouse or rabbit immunoglobulins were used as secondary antibodies (diluted 1:10 000, Sigma-Aldrich). Immunodetection was achieved with a chemiluminescence reagent (Pierce) and detected by LAS 4000 (Fuji Film). For immunoprecipitation, each of the DNA fragments encoding Flag-BAX, Flag-BAX(AAA), Flag-BAX(L63A), HA-A1, Flag-BIMEL, Flag-BAD or Flag-Noxa was cloned into pcDNA5/FRT/TO (Invitrogen) and ectopically expressed in 293T cells. Cells were harvested and then lysed in a 0.5% Triton X-100 buffer (Boston BioProduct) supplemented with a complete protease inhibitor cocktail (Roche). After pre-clearing with protein A/G agarose beads for 1 h at 4 °C, whole-cell lysates were used for immunoprecipitation with antibodies. Generally, 1-4 μg of the commercial antibodies was added to 1 ml of the cell lysate, which was then incubated at 4 °C for 8-12 h. After addition of protein A/G agarose beads, incubation was continued for additional 2 h. Immunoprecipitates were extensively washed with a Triton X-100 lysis buffer and eluted with a SDS-PAGE loading buffer by boiling for 5 min. Immunoprecipitation and immunoblot analyses involved antibodies against BCL-2 (1:100, Santa Cruz Biotechnology), BCL-XL (1:1 000, BD Pharmingen), BCL-w (1:500, BD Pharmingen), MCL-1 (1:100, Santa Cruz Biotechnology), BAX (1:500, BD Transduction Laboratory), BAK (1:500, Sigma-Aldrich), M2 Flag (1:2 000, Sigma-Aldrich), HA (1:1 000, Covance) or actin (1:2 000, Sigma-Aldrich).
Apoptosis analyses
Bax−/− Bak−/− MEF cells stably expressing the wild-type or mutant forms of BAX were seeded at 1 × 106 cells per well into six-well plates for 24 h. The cells were then treated with fresh medium containing 50-100 μM etoposide (Sigma-Aldrich). For the cell viability assay, the cells were stained with propidium iodide (PI) and cells that do not take up PI were quantified using flow cytometry. For the analysis of apoptotic cells, the samples were prepared using a Vybrant Apoptosis Assay kit (Invitrogen) according to the manufacturer's instructions. Nuclei were counterstained with PI. The percentage of annexin V-positive cells was determined by flow cytometry. For the sub-G1 phase PI staining assay, the cells were collected with the cell dissociation buffer (Sigma-Aldrich) and then fixed with 70% ethanol overnight at −20 °C. Fixed cells were washed twice with PBS, and incubated in the PBS buffer containing PI (5 μg/ml), RNase A (1 mg/ml) and Triton X-100 (0.5%) at room temperature for 30 min. Fluorescence emitted from the PI-DNA complex was measured using FACScan flow cytometry. Cells containing hypodiploid DNA were considered apoptotic. The data were analyzed using FlowJo-6.4.
Immunofluorescence and confocal laser scanning microscopy
Bax−/− Bak−/− MEF stable cells grown on eight-well chamber slides were fixed with 2% (w/v) paraformaldehyde in the PBS buffer for 20 min, permeabilized with 0.2% (v/v) Triton X-100 for 15 min and blocked with 10% goat serum (Gibco-BRL) for 1 h. Primary antibody staining was performed using antiserum or purified antibodies in 1% goat serum for 1-2 h at room temperature. The cells were then extensively washed with the PBS buffer and incubated with diluted secondary antibodies in 1% goat serum for 1 h. The cells were mounted using Vectashield (Vector Laboratories, Inc.). The confocal images were acquired using a Nikon Eclipse C1 laser-scanning microscope (Nikon) fitted with a ×100 Nikon objective and Nikon Element image software.
Accession numbers for the protein sequences
The accession numbers in the sequence databases are BCL-2 (NP_000624), BCL-XL (NP_033873), BCL-w (NP_031563), MCL-1 (NP_032588), A1 (NP_033872), BIM (NP_997563.1), BID (NP_031570.2), PUMA (NP_573497.1), BAD (NP_031548.1), BIK (NP_031572.1), BMF (NP_612186.2), Hrk (NP_031571.1), Noxa (NP_067426.1), BAX (NP_031553) and BAK (NP_031549).
Accession number
The coordinates of the BCL-2-BAX peptide structure together with the structure factors have been deposited in the Protein Data Bank with the accession code of 2XA0.
Accession codes
Accessions
GenBank/EMBL/DDBJ
Protein Data Bank
References
Adams JM, Cory S . Life-or-death decisions by the Bcl-2 protein family. Trends Biochem Sci 2001; 26:61–66.
Opferman JT, Korsmeyer SJ . Apoptosis in the development and maintenance of the immune system. Nat Immunol 2003; 4:410–415.
Antignani A, Youle RJ . How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr Opin Cell Biol 2006; 18:685–689.
Green DR, Kroemer G . The pathophysiology of mitochondrial cell death. Science 2004; 305:626–629.
Kim H, Rafiuddin-Shah M, Tu HC, et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol 2006; 8:1348–1358.
Willis SN, Fletcher JI, Kaufmann T, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science 2007; 315:856–859.
Sattler M, Liang H, Nettesheim D, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science 1997; 275:983–986.
Liu X, Dai S, Zhu Y, Marrack P, Kappler JW . The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity 2003; 19:341–352.
Petros AM, Nettesheim DG, Wang Y, et al. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci 2000; 9:2528–2534.
Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ . Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2002; 2:183–192.
Cartron PF, Gallenne T, Bougras G, et al. The first alpha helix of Bax plays a necessary role in its ligand-induced activation by the BH3-only proteins Bid and PUMA. Mol Cell 2004; 16:807–818.
Certo M, Del Gaizo Moore V, Nishino M, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 2006; 9:351–365.
Kim H, Tu HC, Ren D, et al. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell 2009; 36:487–499.
Adams JM, Cory S . Bcl-2-regulated apoptosis: mechanism and therapeutic potential. Curr Opin Immunol 2007; 19:488–496.
Uren RT, Dewson G, Chen L, et al. Mitochondrial permeabilization relies on BH3 ligands engaging multiple prosurvival Bcl-2 relatives, not Bak. J Cell Biol 2007; 177:277–287.
Fletcher JI, Meusburger S, Hawkins CJ, et al. Apoptosis is triggered when prosurvival Bcl-2 proteins cannot restrain Bax. Proc Natl Acad Sci USA 2008; 105:18081–18087.
Youle RJ, Strasser A . The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 2008; 9:47–59.
Zhai D, Jin C, Huang Z, Satterthwait AC, Reed JC . Differential regulation of Bax and Bak by anti-apoptotic Bcl-2 family proteins Bcl-B and Mcl-1. J Biol Chem 2008; 283:9580–9586.
Liu D, Yang B, Cao R, Huang Z . A chemical strategy to promote helical peptide-protein interactions involved in apoptosis. Bioorg Med Chem Lett 2005; 15:4467–4469.
Chen L, Willis SN, Wei A, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 2005; 17:393–403.
Ku B, Woo JS, Liang C, et al. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog 2008; 4:e25.
Willis SN, Chen L, Dewson G, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev 2005; 19:1294–1305.
Hsu YT, Youle RJ . Bax in murine thymus is a soluble monomeric protein that displays differential detergent-induced conformations. J Biol Chem 1998; 273:10777–10783.
Simmons MJ, Fan G, Zong WX, Degenhardt K, White E, Gelinas C . Bfl-1/A1 functions, similar to Mcl-1, as a selective tBid and Bak antagonist. Oncogene 2008; 27:1421–1428.
Werner AB, de Vries E, Tait SW, Bontjer I, Borst J . Bcl-2 family member Bfl-1/A1 sequesters truncated bid to inhibit is collaboration with pro-apoptotic Bak or Bax. J Biol Chem 2002; 277:22781–22788.
Zhang H, Cowan-Jacob SW, Simonen M, Greenhalf W, Heim J, Meyhack B . Structural basis of BFL-1 for its interaction with BAX and its anti-apoptotic action in mammalian and yeast cells. J Biol Chem 2000; 275:11092–11099.
Suzuki M, Youle RJ, Tjandra N . Structure of Bax: coregulation of dimer formation and intracellular localization. Cell 2000; 103:645–654.
Hsu YT, Youle RJ . Nonionic detergents induce dimerization among members of the Bcl-2 family. J Biol Chem 1997; 272:13829–13834.
Oltvai ZN, Milliman CL, Korsmeyer SJ . Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 1993; 74:609–619.
Zha H, Aime-Sempe C, Sato T, Reed JC . Proapoptotic protein Bax heterodimerizes with Bcl-2 and homodimerizes with Bax via a novel domain (BH3) distinct from BH1 and BH2. J Biol Chem 1996; 271:7440–7444.
Moldoveanu T, Liu Q, Tocilj A, Watson M, Shore G, Gehring K . The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol Cell 2006; 24:677–688.
Leber B, Lin J, Andrews DW . Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis 2007; 12:897–911.
Cuconati A, Mukherjee C, Perez D, White E . DNA damage response and MCL-1 destruction initiate apoptosis in adenovirus-infected cells. Genes Dev 2003; 17:2922–2932.
Chittenden T, Flemington C, Houghton AB, et al. A conserved domain in Bak, distinct from BH1 and BH2, mediates cell death and protein binding functions. EMBO J 1995; 14:5589–5596.
Tan C, Dlugosz PJ, Peng J, et al. Auto-activation of the apoptosis protein Bax increases mitochondrial membrane permeability and is inhibited by Bcl-2. J Biol Chem 2006; 281:14764–14775.
Zhou H, Hou Q, Hansen JL, Hsu YT . Complete activation of Bax by a single site mutation. Oncogene 2007; 26:7092–7102.
Gavathiotis E, Suzuki M, Davis ML, et al. BAX activation is initiated at a novel interaction site. Nature 2008; 455:1076–1081.
Walensky LD, Pitter K, Morash J, et al. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell 2006; 24:199–210.
Lovell JF, Billen LP, Bindner S, et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell 2008; 135:1074–1084.
Petros AM, Medek A, Nettesheim DG, et al. Solution structure of the antiapoptotic protein bcl-2. Proc Natl Acad Sci USA 2001; 98:3012–3017.
Smits C, Czabotar PE, Hinds MG, Day CL . Structural plasticity underpins promiscuous binding of the prosurvival protein A1. Structure 2008; 16:818–829.
Vagin A, Teplyakov A . MOLREP: an Automated Program for Molecular Replacement. J Appl Cryst 1997; 30:1022–1025.
Emsley P, Cowtan K . Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126–2132.
Brunger AT, Adams PD, Clore GM, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 1998; 54:905–921.
Acknowledgements
This study made use of the beam line 4A at the Pohang Accelerator Laboratory in Korea. This work was supported by the GRL Program (K20815000001) from the National Research Foundation of Korea (B-HO and JUJ); the Brain Korea 21 Project (BK); US Public Health Service grants CA140964, AI083841; the Leukemia & Lymphoma Society of USA; and the Wright Foundation and the Baxter Foundation (CL).
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Table S1
Data collection and structure refinement statistics (PDF 59 kb)
Supplementary information, Figure S1
Comparison of the structures of antiapoptotic BCL-2 proteins bound to a BH3 peptide (PDF 251 kb)
Supplementary information, Figure S2
The BH3 domain-containing segment in the structure of free BAX (PDF 146 kb)
Supplementary information, Figure S3
Phenotypes of BAX(AAA) (PDF 139 kb)
Supplementary information, Figure S4
Expression levels of proteins (PDF 108 kb)
Rights and permissions
About this article
Cite this article
Ku, B., Liang, C., Jung, J. et al. Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res 21, 627–641 (2011). https://doi.org/10.1038/cr.2010.149
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2010.149
Keywords
This article is cited by
-
Parkin-mediated ubiquitination inhibits BAK apoptotic activity by blocking its canonical hydrophobic groove
Communications Biology (2023)
-
Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity
Nature Communications (2023)
-
Neuroprotective effect of a novel brain-derived peptide, HIBDAP, against oxygen-glucose deprivation through inhibition of apoptosis in PC12 cells
Molecular Biology Reports (2023)
-
Multi-catalytic Sites Inhibition of Bcl2 Induces Expanding of Hydrophobic Groove: A New Avenue Towards Waldenström Macroglobulinemia Therapy
The Protein Journal (2022)
-
Ethyl acetate extract of Elephantopus mollis Kunth induces apoptosis in human gastric cancer cells
BMC Complementary Medicine and Therapies (2021)