Abstract
Glial cell line-derived neurotrophic factor (GDNF) was originally recognized for its ability to promote survival of midbrain dopaminergic neurons, but it has since been demonstrated to be crucial for the survival and differentiation of many neuronal subpopulations, including motor neurons, sympathetic neurons, sensory neurons and enteric neurons. To identify possible effectors or regulators of GDNF signaling, we performed a yeast two-hybrid screen using the intracellular domain of RET, the common signaling receptor of the GDNF family, as bait. Using this approach, we identified Rap1GAP, a GTPase-activating protein (GAP) for Rap1, as a novel RET-binding protein. Endogenous Rap1GAP co-immunoprecipitated with RET in neural tissues, and RET and Rap1GAP were co-expressed in dopaminergic neurons of the mesencephalon. In addition, overexpression of Rap1GAP attenuated GDNF-induced neurite outgrowth, whereas suppressing the expression of endogenous Rap1GAP by RNAi enhanced neurite outgrowth. Furthermore, using co-immunoprecipitation analyses, we found that the interaction between RET and Rap1GAP was enhanced following GDNF treatment. Mutagenesis analysis revealed that Tyr981 in the intracellular domain of RET was crucial for the interaction with Rap1GAP. Moreover, we found that Rap1GAP negatively regulated GNDF-induced ERK activation and neurite outgrowth. Taken together, our results suggest the involvement of a novel interaction of RET with Rap1GAP in the regulation of GDNF-mediated neurite outgrowth.
Similar content being viewed by others
Introduction
Glial cell line-derived neurotrophic factor (GDNF) was originally discovered by its ability to promote the survival of midbrain dopaminergic neurons 1 but has since been demonstrated to be crucial for the survival and differentiation of many neuronal subpopulations, including motor neurons, sympathetic neurons, sensory neurons and enteric neurons 2, 3, 4. Moreover, GDNF is required to induce branching of ureteric buds during kidney development 5 and is important for cell fate decision of undifferentiated spermatogonia in the testis 6. The significance of GDNF is underscored by its potent neuroprotective and restorative effects in several animal models of Parkinson's disease and motor neuron disease, making it an attractive therapeutic candidate for treatment of neurodegenerative diseases 7, 8.
GDNF signals through a multi-component receptor system consisting of the GDNF family receptor-α and the RET receptor tyrosine kinase or NCAM 9, 10. All members of the GDNF family, including GDNF, neurturin, artemin and persephin, share RET as their common signaling receptor, and their binding specificity is determined by distinct GDNF receptor α proteins 3. Activation of the receptor tyrosine kinase RET, which can be achieved either by interactions between GDNF receptor-α subunits and GDNF family members or by various oncogenic mutations, results in distinct cellular outcomes, including proliferation, differentiation, survival and motility 3, 4. Ablation of RET results in defects in the kidney and enteric nervous system in mice 11. In addition, RET-deficient mice show progressive, adult-onset loss of dopaminergic neurons in the nigrostriatal system 12. In humans, inactivating mutations in RET lead to the development of Hirshsprung's disease, which is characterized by megacolon aganglionosis 13, 14, while activating mutations cause various types of endocrine tumors 15. Several experiments have shown that upon activation, RET recruits various signaling modules and triggers a variety of intracellular signaling pathways, including the Ras or Rap1/ERK, phosphatidylinositol 3-kinase (PI3K)/Akt, phospholipase Cγ (PLCγ) and Rac/c-Jun N-terminal kinase (JNK) pathways 16, 17, 18, 19, 20, 21, 22. However, despite considerable progress in understanding the RET signaling pathways responsible for cell survival and proliferation, the mechanism underlying RET-mediated neuronal differentiation remains largely unknown.
To gain insight into the mechanisms by which GDNF-mediated activation of RET enhances neuronal differentiation, we performed a yeast two-hybrid screen on a human brain cDNA library using the intracellular domain of RET as bait, and we identified Rap1GAP as a candidate binding partner for RET. Rap1GAP belongs to the family of GTPase activating proteins (GAPs), which accelerate hydrolysis of bound GTP to GDP, to block the activity of small G proteins. Specifically, Rap1GAP suppresses the activation of Rap1, a small G protein with 53% amino acid identity to Ras 23, 24. Members of the Rap1GAP family (Rap1GAPs), including Rap1GAP, Rap1GAPII, signal-induced proliferation-associated gene-1 (SPA-1), spine-associated RapGAP (SPAR), E6-targeted protein 1 (E6TP1) and several SPA-1-like proteins (SPA-Ls), are highly expressed in non-proliferating tissues, such as the nervous system and pancreas 25. In addition, Rap1GAPs act as tumor suppressors. For example, degradation of the E6TP1 protein increases the incidence of cervical cancer 26, while deletion of the spa1 gene in mice creates a spectrum of myeloid disorders that resemble chronic myeloid leukemia 25. Furthermore, SPAR is involved in regulating synaptic plasticity and enlargement of dendrite spine heads 27. However, the roles of Rap1GAP in the nervous system are largely unknown. Thus far, there is no evidence to show that Rap1GAPs interact with any receptor tyrosine kinases (RTKs) that play a crucial role in neural development.
In the present study, we identified a novel interaction between RET and Rap1GAP and demonstrated that Rap1GAP negatively regulates GNDF-induced ERK activation and neurite outgrowth. Our results suggest a novel role for Rap1GAP in the regulation of GDNF-mediated cell differentiation.
Results
Rap1GAP is identified as a RET-binding protein in yeast-hybrid screen
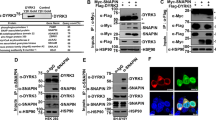
To identify potential interaction partners for RET within the nervous system, we performed a yeast two-hybrid screening on a human brain cDNA library using the intracellular domain of RET (amino acids 658-1 114) as bait (Figure 1A). Among the positive clones, we identified one as a cDNA fragment corresponding to amino-acid residues 26-223 of Rap1GAP. This region includes a portion of the GAP catalytic domain and the N-terminal flanking sequences, as shown in Figure 1B. We used a β-galactosidase activity filter assay and liquid culture assay to further test the interaction between full-length Rap1GAP and RET. As shown in Figure 1C and 1D, RET showed a robust interaction with Rap1GAP, while the EGF receptor, another RTK, did not. Thus, according to our yeast two-hybrid assays, Rap1GAP selectively interacts with the intracellular domain of RET.
RET interacts with Rap1GAP in a yeast two-hybrid screen. (A) Schematic representation of the pGilda-RETIC bait. Cad, Cadherin domain; Cys, cysteine-rich domain; TM, transmembrane domain; TK, tyrosine kinase domain; LexA, LexA fusion vector pGilda. (B) A fragment of human Rap1GAP (amino acids 26-223) was isolated from the two-hybrid screen. GAP, GTPase-activating protein domain. (C, D) A filter assay and (C) liquid culture assay using o-nitrophenyl-D-galactoside (ONPG) (D) were performed to analyze β-galactosidase activity. Full-length wild-type Rap1GAP was co-transformed into the yeast reporter strain EGY48 along with the bait encoding the intracellular domain of RET or the EGF receptor. Positive and negative controls are described in the Materials and Methods section.
RET immunoprecipitates with Rap1GAP in neural tissues
To test whether endogenous Rap1GAP and RET could form a complex in mammalian tissues, we performed immunoprecipitation assays with homogenates of rat neural tissues. We first examined the expression of the two proteins in various tissues. As shown in Figure 2A, Rap1GAP was expressed in the midbrain, spinal cord, basal forebrain, hippocampus, hypothalamus and cerebellum, while RET was expressed in the midbrain, spinal cord, basal forebrain, hypothalamus and cerebellum but not in the hippocampus. These results are consistent with previous reports 28. In NIH3T3 cells, neither RET nor Rap1GAP was detectable, while in PC12 cells, only Rap1GAP was expressed. Proteins from these homogenates were immunoprecipitated with an anti-RET antibody and immunoblotted with anti-Rap1GAP or anti-RET antibodies. As shown in Figure 2B, Rap1GAP co-immunoprecipitated with RET in the midbrain and spinal cord, but not in hippocampus due to the lack of RET in hippocampus. In addition, the homogenate proteins were immunoprecipitated with an anti-Rap1GAP antibody and immunoblotted with anti-RET or anti-Rap1GAP antibodies. RET co-immunoprecipitated with Rap1GAP in midbrain and spinal cord but not in the hippocampus, while Rap1GAP was expressed in all of these tissues. These results were further confirmed by using control IgG immunoprecipitation in rat midbrain tissue (Figure 2C). Thus, endogenous RET and Rap1GAP could interact with one another, providing a basis for their physiological function.
Endogenous Rap1GAP interacts with RET in the rat mesencephalon and spinal cord. (A) Western blot analysis of Rap1GAP and RET in rat neural tissues and cell lines. (B) Co-immunoprecipitation of endogenous RET with Rap1GAP. Tissue homogenates from the midbrain, spinal cord and hippocampus as well as from NIH3T3 cells were prepared, immunoprecipitated with an anti-RET or anti-Rap1GAP antibody and then immunoblotted with an anti-Rap1GAP or anti-RET antibody, respectively. (C) Co-immunoprecipitation of endogenous RET, with Rap1GAP or Rap1GAP, with RET from rat midbrain tissues. Rabbit IgG was used as an immunoprecipitation control, and the heavy chain of IgG was visualized for input control. IP, immunoprecipitation; IB, immunoblot.
RET is co-expressed with Rap1GAP in neurons
To obtain further morphological evidence that RET and Rap1GAP are co-expressed in the neural cells, we next carried out immunohistochemical analyses. Specifically, we processed slices of rat mesencephalon for triple labeling immunofluorescence. As shown in Figure 3A, both RET (green) and Rap1GAP (red) were detected in the substantia nigra. Most of the RET-expressing neurons were also immunopositive for the dopaminergic cell marker tyrosine hydroxylase (TH), while Rap1GAP expression was widespread in the substantia nigra. Furthermore, Rap1GAP and RET were co-expressed in TH-positive neurons. In addition, Rap1GAP and RET were co-expressed in spinal cord neurons (data not shown). To clarify the subcellular localization of Rap1GAP and RET, we fixed the primary cultured mesencephalic neurons on coverslips and processed them for immunofluorescence triple labeling. As shown in Figure 3B, RET (green) and Rap1GAP (red) were co-expressed in cultured TH-positive mesencephalic dopaminergic neurons (blue), including both somatodendritic and axonal regions. Furthermore, in the midbrain, we observed that RET (green) and Rap1GAP (red) were co-localized with EEA1 (blue), a specific marker of early endosomes. Thus, these results demonstrate that RET and Rap1GAP are co-expressed in neurons.
Rap1GAP and RET are co-expressed in rat mesencephalic neurons. (A) Tissue slices from the rat mesencephalon were triple labeled with anti-Rap1GAP (red), anti-RET (green) and anti-TH (purple), and protein expression was observed by immunofluorescence confocal microscopy. The merged images show that Rap1GAP and RET were co-expressed in TH-positive mesencephalic neurons. Scale bars, 20 μm. (B) Triple labeling immunofluorescence showed the subcellular localization of RET (green) and Rap1GAP (red) in primary cultured TH-positive (blue) mesencephalic neurons. Scale bar, 5 μm. (C) Triple labeling immunofluorescence showed the subcellular localization of RET (green), Rap1GAP (red) and EEA1 (an early endosome marker, blue) in rat mesencephalic neurons. The merged images demonstrate that Rap1GAP and RET were co-localized with early endosomes. Scale bar, 5 μm.
Rap1GAP suppresses GDNF-induced neurite outgrowth in PC12-GFRα1-RET cells
To identify a possible physiological role of Rap1GAP in the downstream signaling of RET, we first performed a neurite outgrowth assay in PC12 cells, which are representative of differentiated cells. Endogenous Rap1GAP was expressed in PC12 cells; however, little endogenous RET was detected in these cells (Figure 2A). Thus, we used our previously established PC12-GFRα1-RET cell line that stably expresses GFRα1 and RET 22. To explore the influence of Rap1GAP on GDNF-induced cell differentiation, we generated a GFP-Rap1GAP fusion construct and transfected the construct into PC12-GFRα1-RET cells. After incubation with 50 ng/ml GDNF for 3 days, the control cells, transfected with an enhanced green fluorescent protein (EGFP) vector, showed enlarged cell bodies and elongated neurites, suggesting that GDNF stimulates differentiation of PC12-GFRα1-RET cells (Figure 4A). Interestingly, overexpression of GFP-Rap1GAP significantly attenuated the neurite outgrowth induced by GDNF (Figure 4A and 4B), indicating that Rap1GAP negatively regulates the signaling activated by GDNF that mediates neurite outgrowth.
Rap1GAP negatively regulates GDNF-induced neurite outgrowth in PC12-GFRα1-RET cells. (A) PC12-GFRα1-RET cells were transfected with the pEGFP-Rap1GAP or EGFP vector and then incubated with or without GDNF for 72 h. Cells were visualized using fluorescence microscope (with a 20× objective) based on GFP expression. (B) Quantification (shown as mean ± s.d.) of neuronal differentiation of PC12-GFRα1-RET cells overexpressing Rap1GAP. **P < 0.01 as indicated. (C) Western blot analysis was used to evaluate Rap1GAP expression in cells 72 h after transfection with various siRNA constructs. All the cell extracts were immunoblotted with a Rap1GAP antibody. Lane 1, missense RNAi was used as a negative control; lanes 2-4, various siRNA constructs against Rap1GAP. Anti-actin was used as a loading control. (D) PC12-GFRα1-RET cells transfected with Rap1GAP siRNA or control siRNA were serum starved and then incubated with or without GDNF for 72 h. Cells were visualized using fluorescence microscope. (E) Quantification (shown as mean ± s.d.) of the neuronal differentiation of PC12-GFRα1-RET cells following Rap1GAP RNAi treatment. **P < 0.01 as indicated. (F) Rap1GAP interacts with RET in PC12-GFRα1-RET cells. PC12-GFRα1-RET cells were serum-starved for 12 h and then treated with or without 50 ng/ml GDNF for 10 min. The cell lysates were immunoprecipitated with anti-RET or anti-Rap1GAP followed by immunoblotting with anti-Rap1GAP or anti-RET, respectively. Immunoprecipitation with normal IgG was used as a negative control. The inputs represent 5% of the lysates used for the immunoprecipitation reaction. The data are representative of four independent experiments. IP, immunoprecipitation; IB, immunoblot. (G) Quantification of Rap1GAP bound to RET in PC12-GFRα1-RET cells shown in Figure 4F. Blots of Rap1GAP or RET without GDNF treatment were used for normalization. The data represent the mean ± s.d. of four independent experiments. *P < 0.05.
To further support the effect of Rap1GAP on GDNF-induced cell differentiation, we employed an RNAi approach. We designed and synthesized three siRNA sequences targeting Rap1GAP. Western blot analyses showed that all these sequences were able to knock down Rap1GAP expression levels effectively (Figure 4C). We selected the most effective RNAi sequence and co-transfected it with GFP into PC12-GFRα1-RET cells. We then examined the differentiation of GFP-positive cells following GDNF stimulation. Compared with the RNAi control group, suppressing Rap1GAP gene expression by RNAi significantly increased GDNF-induced cell differentiation (Figure 4D and 4E). These results further suggest that endogenous Rap1GAP is a crucial negative regulator in GDNF-induced cell differentiation.
To determine whether endogenous Rap1GAP associates with RET in PC12-GFRα1-RET cells, we performed co-immunoprecipitation assays. We immunoprecipitated cell lysates with either an anti-RET or anti-Rap1GAP antibody, then immunoblotted with an anti-Rap1GAP or anti-RET antibody, respectively. As shown in Figure 4F and 4G, we detected an interaction between Rap1GAP and RET in lysates from cells that were treated with or without GDNF, and the interaction was strengthened following GDNF treatment. Under the same conditions, RET and Rap1GAP did not co-immunoprecipitate when control IgG was used. These results demonstrate that GDNF treatment significantly enhanced the RET-Rap1GAP interaction in PC12-GFRα1-RET cells, strongly suggesting that Rap1GAP is involved in GDNF signaling.
Identification of the crucial binding site for the interaction of RET with Rap1GAP
GNDF treatment significantly enhanced the RET-Rap1GAP interaction in PC12-GFRα1-RET cells, suggesting that this interaction is ligand dependent. RET exerts its biological effects mainly through distinct autophosphorylation of tyrosine residues. Five putative tyrosine phosphorylation sites within the intracellular domain of RET (Y905, Y981, Y1015, Y1062 and Y1096) serve as potential docking sites for signaling molecules 29. To identify which residue(s) may be involved in the interaction with Rap1GAP, we constructed five RET mutants (Y905F, Y981F, Y1015F, Y1062F and Y1096F) by mutating tyrosine residues to phenylalanine, as done in our previous study 22. We also constructed RET (K758M), which changes Lys758 of the ATP-binding site within the kinase domain of RET to Met, rendering the receptor catalytically inactive 30. We then co-transfected these mutant constructs into HEK293T cells together with Rap1GAP and GFRα1 and stimulated the cells with GDNF. The K758M mutant of RET (i.e., the kinase-dead mutant) almost abolished the interaction with Rap1GAP (Figure 5A). The mutation of Tyr981 to Phe significantly attenuated the interaction between RET and Rap1GAP, whereas none of the other four tyrosine mutations appeared to be relevant to Rap1GAP binding because they did not obviously impair the interaction (Figure 5A). These results suggest that Y981 is crucial to the interaction of RET with Rap1GAP. To further identify whether Rap1GAP interacts with phosphorylated RET, we co-transfected wild-type RET and RET (K758M) into HEK293T cells together with Rap1GAP and GFRα1, and then stimulated the cells with GDNF. Next, we immunoprecipitated cell lysates with an anti-Rap1GAP antibody and immunoblotted with an antibody against phosphorylated RET. As shown in Figure 5B, we found that Rap1GAP interacted with phosphorylated RET.
Identification of the binding site for Rap1GAP in RET. (A) HEK293T cells were co-transfected with wild-type RET or the indicated RET mutants along with Rap1GAP and GFRα1, and cells were then stimulated with 50 ng/ml GDNF for 10 min. Cell extracts were analyzed by immunoprecipitation (IP) with anti-RET or anti-Rap1GAP followed by immunoblotting (IB) with antibodies against Rap1GAP or RET, respectively. The bottom two panels show RET or Rap1GAP expression in total extracts. WT, wild type; K, lysine; M, methionine; Y, tyrosine; F, phenylalanine. (B) HEK293T cells were co-transfected with wild-type RET or the RET (K758M) mutant, along with Rap1GAP and GFRα1, and cells were then stimulated with 50 ng/ml GDNF for 10 min. Cell extracts were analyzed by immunoprecipitation (IP) with anti-Rap1GAP followed by immunoblotting (IB) with antibodies against Rap1GAP, RET or phospho-RET (905).
Rap1GAP suppresses GDNF-induced neurite outgrowth by inhibiting ERK activation
Differentiation of PC12 cells is mainly associated with the activation of the Ras-Raf-MEK-ERK and PI3K-Akt signaling pathways 31, 32. Therefore, using selective kinase inhibitors, we examined whether blockade of each of these pathways could inhibit GDNF-induced neurite outgrowth. As shown in Figure 6A, blockade of the ERK pathway by the MEK1 inhibitor PD98059 significantly inhibited GDNF-induced neurite outgrowth in PC12-GFRα1-RET cells, whereas the PI-3 kinase inhibitor LY294002 had little effect. These results suggest that GDNF-induced neurite outgrowth is highly dependent on the ERK pathway but not on the PI-3 kinase pathway. To further examine the effects of Rap1-GAP on ERK activation, we overexpressed or knocked down Rap1GAP by transfection with GFP-Rap1GAP or Rap1GAP siRNA, respectively, in PC12-GFRα1-RET cells. After the cells were treated with GDNF, we collected the lysates and subjected them to western blot analysis with an anti-pERK antibody. As shown in Figure 6B and 6C, GDNF induced sustained ERK phosphorylation, and this effect was strengthened by siRNA treatment against Rap1GAP. In contrast, the pERK level was suppressed by overexpression of Rap1GAP, indicating an inhibitory effect of Rap1GAP on ERK signaling. We also performed pull-down assays to detect Rap1 activation using the GST-tagged Rap1GTP-binding domain of RalGDS (GST-RalGDS-RBD) to capture Rap1 in the GTP-bound state. As shown in Figure 6B and 6D, overexpression of Rap1GAP greatly attenuated the Rap1GTP level, whereas knockdown of Rap1GAP increased Rap1GTP level. Taken together, these results suggest that Rap1GAP negatively regulates GDNF-induced neurite outgrowth by inhibiting ERK activation.
Rap1GAP inhibits GDNF-induced activation of the ERK signaling pathway. (A) Effects of kinase inhibitors on GDNF-induced differentiation of PC12-GFRα1-RET cells. PC12-GFRα1-RET cells were serum starved and incubated for 72 h with or without 50 ng/ml GDNF in the presence of DMSO as a control, PD98059 (10 μM), or LY294002 (10 μM). Data are shown as the mean ± s.d. ##P < 0.01 as indicated. (B) PC12-GFRα1-RET cells were transfected with GFP, GFP-Rap1GAP or Rap1GAP siRNA, then serum starved for 6 h, and then stimulated with 50 ng/ml GDNF for the indicated time. Cell extracts were used for western blot analysis with anti-ERK or anti-pERK or were subjected to a Rap1 activity assay, as described in the Materials and Methods section. (C-D) Quantification of relative pERK levels (C) or relative Rap1 activity levels (D). Cells transfected with GFP but not stimulated with GDNF were used for normalization. Data represent the mean ± s.d. of three independent experiments. *P < 0.05; **P < 0.01 as indicated.
Discussion
Given the functional diversity of the RET receptor tyrosine kinase, understanding the complex intracellular signaling pathways triggered by RET activation is important. To gain a better understanding of RET signaling, we sought to identify new RET-interacting proteins. Therefore, we conducted a yeast two-hybrid screen using the intracellular domain of RET as bait, and we identified Rap1GAP as a binding candidate for RET. Rap1GAP belongs to the family of GTPase-activating proteins, which accelerate hydrolysis of bound GTP to GDP, thereby inhibiting the activity of small G proteins, such as Rap1. Thus, Rap1GAP regulates the involvement of these small G proteins in signaling pathways 33. The Gαo activates Rap1 by sequestering Rap1GAP 34, while Rap1GAPII interacts with the Gαi to inactivate Rap1 35, 36, suggesting distinct mechanisms of the Rap1GAPs in regulating G protein-coupled receptor signal transduction. RasGAP has been reported to interact with RTKs (e.g., TrkA and the EGF receptor) to inhibit the transient activation of ERK and to negatively regulate cell proliferation 37, 38, 39. In contrast, Rap1GAP is unable to be recruited to activated TrkA and remains disseminated in the cytoplasm 40. Furthermore, to date, there is no evidence to show that Rap1GAPs interact with RTKs. In the present study, a yeast two-hybrid screen identified a robust interaction between RET and Rap1GAP, while the EGF receptor was not found to bind to Rap1GAP, suggesting a selective interaction between Rap1GAP and RTKs. Furthermore, endogenous Rap1GAP was found to co-immunoprecipitate with RET in neural tissues (e.g., spinal cord and mesencephalon). Morphological studies revealed that RET and Rap1GAP were co-expressed in dopaminergic neurons in the mesencephalon. Moreover, endogenous Rap1GAP interacted with RET in PC12-GFRα1-RET cells, and this interaction was strengthened by GDNF treatment. Mutagenesis analysis demonstrated that Tyr981 within the intracellular domain of RET was crucial for the interaction with Rap1GAP. Thus, this study provides the first evidence that Rap1GAP interacts with RET, suggesting that Rap1GAP may serve as a novel regulator of distinct RTK signaling.
Previously, GDNF was demonstrated to promote neurite outgrowth in various primary cultures of sympathetic neurons 41, midbrain dopaminergic neurons 42, motoneurons 43, sensory neurons 44 and enteric neurons 45, and similar effects have been observed in vivo 46, 47, 48. To explore the function of Rap1GAP in GDNF/RET signaling, we initially chose PC12-GFRα1-RET cells as an experimental model 22, taking advantage of the fact that these cells show neurite outgrowth in response to GDNF stimulation and express endogenous Rap1GAP. Overexpression of GFP-Rap1GAP significantly attenuated GDNF-induced neurite outgrowth, while knockdown of endogenous Rap1GAP by RNAi greatly enhanced GDNF-induced neurite outgrowth. Thus, Rap1GAP is a crucial negative regulator of GDNF-induced neurite outgrowth.
GDNF can trigger many intracellular signaling pathways, including the Ras/ERK, PI3K/Akt and Rac/JNK pathways 3. Among these, the Ras/ERK and PI3K/AKT pathways have been reported to be more relevant to cell differentiation. PI3K/AKT signaling regulates growth cone turning and terminal branching responses, whereas Ras/ERK signaling regulates local signaling related to microtubule assembly and axon extension 49, 50. Recently, RET activation was shown to induce sustained ERK activation via the Rap1 pathway in the TGW neuroblastoma cell line 21. In the present study, GDNF induced sustained activation of Rap1 and ERK in PC12 cells. Our pharmacological data from using selective kinase inhibitors suggested that GDNF-induced neurite outgrowth was highly dependent on the ERK pathway, but not on the PI3K pathway. More importantly, levels of pERK and GTP-Rap1 were suppressed by Rap1GAP overexpression and enhanced by RNAi knockdown of Rap1GAP, indicating an inhibitory effect of Rap1GAP on Rap1-ERK signaling. Thus, Rap1GAP may negatively regulate GDNF-induced neurite outgrowth by inhibiting ERK activation.
Materials and Methods
Plasmid constructs
Full-length RET constructs were a generous gift from Dr Carlos F Ibanez. The intracellular domain of human RET (amino acids 658-1 114) was generated by PCR and cloned in-frame into the LexA fusion vector pGilda (Clontech) to generate the bait pGilda-RETIC. Full-length Rap1GAP cDNA was a generous gift from Dr Patrick J Casey. Rap1GAP was cloned into pB42AD and EGFPN1 vectors. All constructs were fully sequenced before being used for transformation or transfection.
Yeast two-hybrid screen and assay
The yeast two-hybrid screen of a human brain cDNA library was performed as previously described 22. Filter-lift color assays and liquid culture assays with o-nitrophenyl-D-galactoside (ONPG) for β-galactosidase activity analysis were also performed as described previously 51. During the analysis, pGilda-53 co-transformed with pB42AD-T was used as a positive control, and pGilda co-transformed with pB42AD was used as a negative control.
Cell culture and transfection
Wild-type PC12 cells and PC12-GFRα1-RET cells 22 were grown in Dulbecco's modified Eagle's medium (DMEM, Gibco BRL) supplemented with 5% fetal bovine serum (FBS) and 5% heat-inactivated horse serum (HyClone). Hygromycin and G418 were used to maintain exogenous protein expression in PC12-GFRα1-RET cells. Lipofectamine (Invitrogen) and a calcium phosphate reagent (Promega) were used to transfect PC12-GFRα1-RET cells with the pEGFP-Rap1GAP plasmid or vector alone according to the manufacturer's instructions. For immunoprecipitation and immunoblotting assays, 24 h after transfection, cells were starved for 12 h or 6 h and subsequently stimulated with 50 ng/ml GDNF for 10 min at 37 °C.
RNAi
Three target siRNAs against full-length Rap1GAP were designed (no. 1, 715-735, 5′-CTGGTCTTCTCGCTCAAGTAT-3′; no. 2, 1 004-1 024, 5′-TGTCCACCAAGCTGCCATACA-3′; no. 3, 2 908-2 928, 5′-TTGTCCTAGGACCGCCTATCA-3′) and the oligonucleotides were chemically synthesized by GenePharma (Shanghai, China). The scrambled sequence of the control siRNA was 5′-GTTCTCCGAACGTGTCACGT-3′. PC12-GFRа1-RET cells were seeded in 24-well plates (2 × 105 per well) overnight and then transfected with Rap1GAP siRNA (20 μM) using 4 μl of Lipofectamine 2000 (Invitrogen) in 0.5 ml of DMEM containing 10% FBS. The transfection medium was replaced with fresh culture medium 6 h later. The knockdown efficiency of each siRNA construct against Rap1GAP was determined by western blot analysis 72 h after transfection. The no. 2 siRNA oligo was used for the PC12 differentiation and RET internalization assays. This sequence was synthesized and subcloned into the U6/GFP/Neo shRNA vector by GenePharma, and the U6-GFP-Rap1GAP RNAi construct was used in the neurite outgrowth assay by transfection with a calcium phosphate reagent (Promega).
Immunoprecipitation and immunoblotting
Various neural tissues were dissected and isolated from adult male Sprague Dawley rats. Each tissue was homogenized using a pestle tissue grinder at slow speed in solubilization buffer (25 mM HEPES-NaOH (pH 7.4), 125 mM potassium acetate, 5 mM MgCl2, 0.32 M sucrose and 1% Triton X-100). Proteins solubilized from each rat tissue homogenate were collected.
The harvested cells were washed twice with cold phosphate-buffered saline (PBS), solubilized with ice-cold lysis buffer (50 mM Tris-HCl, pH 8.0, 1 mM EDTA, 150 mM NaCl, 0.5% sodium deoxycholate, 0.02% sodium azide, 1 mM NaF, 1 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride, 1% Nonidet P-40, 1 mM dithiothreitol, 0.1% sodium dodecyl sulfate (SDS), 2 μg/ml pepstatin, 2 μg/ml leupeptin and 2 μg/ml aprotinin) and incubated on ice for 20 min. Lysates were clarified by centrifugation at 11 200× g for 10 min at 4 °C.
Protein concentration of the supernatants from tissue homogenates or cell lysates was determined by the Bradford method. For immunoprecipitation, 500 μl of the supernatant was incubated with 5 μl of the precipitating antibody for 3 h at 4 °C under mild agitation. Protein G-agarose beads (Roche) were then added and incubated for 3 h. The immunoprecipitated samples were washed three times with lysis buffer, boiled for 3-5 min in sample-loading buffer, subjected to SDS polyacrylamide gel electrophoresis (SDS-PAGE), immunoblotted with corresponding antibodies and visualized with enhanced chemiluminescence (ECL, Pierce). The antibodies against Rap1GAP (sc-10330) 34, RET (sc-1290) 52, ERK(sc-94) 22 and pERK (sc-7383) 22 were from Santa Cruz; anti-actin was from Kangcheng and anti-phospho-RET (905) was from Cell Signaling 53.
Neurite outgrowth assays
Neurite outgrowth assays were performed as previously described 22. Briefly, PC12-GFRα1-RET cells were seeded in 12-well plates coated with poly-L-lysine at a density of 1 × 105 cells/ml. The cultures were kept in an atmosphere of 5% CO2 and 95% air at 37 °C, and the medium was changed every 3 days. After transfection, the cells were washed twice with PBS (pH 7.4) and then supplied with new medium supplemented with or without 50 ng/ml GDNF. GFP-positive cells were observed under a fluorescence microscope (Olympus, excitation at 454 nm) 3 days later. Cells with one or more neurites of a length more than twice the diameter of the cell body were scored as neurite-bearing positive cells. At least 100 cells per group were randomly counted for each experiment, and the experiment was repeated five times.
Immunohistochemistry and immunocytochemistry
Animal experiments were carried out in accordance with NIH guidelines. Five adult male Sprague Dawley rats were anesthetized with sodium pentobarbital (50 mg/kg, i.p.) and perfused transcardially with 0.1 M PB (pH 7.4), followed by perfusion with a solution of 4% paraformaldehyde and 0.2% saturated picric acid in 0.1 M PB. The brains were removed, kept for 1.5 h in the same fixative at 4 °C and cryoprotected overnight at 4 °C in 0.01 M PBS (pH 7.4) with 20% sucrose. The tissues were then sliced into 14-μm sections on a Leica 1900 cryostat and mounted onto glass slides. The sections were washed in PBS, incubated with the primary antibodies (goat anti-RET 54, Neuromics; rabbit anti-Rap1GAP55, Epitomics; mouse anti-TH 22, Sigma; or mouse anti-EEA1, Chemicon) in PBS containing 3% bovine serum albumin (BSA) and 0.3% Triton X-100 at 4 °C for 48 h. After three washes in PBS, the sections were incubated with secondary antibodies from Jackson ImmunoResearch (FITC-conjugated donkey anti-goat, rhodamine-conjugated donkey anti-rabbit or cy5-conjugated donkey anti-mouse). Finally, the sections were washed and examined with a Leica SP2 confocal microscope.
Cultured mesencephalic neurons on cover glasses were fixed in 4% paraformaldehyde with 0.2% saturated picric acid in 0.1 M phosphate buffer (PB) for 15 min at room temperature. After one wash with 0.01 M PBS, cells were incubated with primary antibodies (rabbit anti-RET(sc-13104) 52, Santa Cruz; goat anti-Rap1GAP(sc-10330) 34, Santa Cruz; mouse anti-TH 22, Sigma) in PBS containing 3% BSA and 0.3% Triton X-100 overnight at 4 °C. Following three washes in 0.01 M PBS, the cells were incubated with secondary antibodies from Jackson ImmunoResearch (FITC-conjugated donkey anti-rabbit, rhodamine-conjugated donkey anti-goat or cy5-conjugated donkey anti-mouse) for 30 min at room temperature. Subsequently, the coverslips were washed in 0.01 M PBS and examined with a Leica SP2 confocal microscope.
Rap1 activity assay
GDNF-treated or untreated cells were lysed on ice for 10 min in lysis buffer (50 mM Tris-HCl (pH 7.4), 0.5 M NaCl, 1% NP40, 2.5 mM MgCl2, 10% glycerol, 1 mM sodium orthovanadate, 250 mM PMSF, 10 μg/ml aprotinin and 10 μg/ml leupeptin) and the resulting lysates were centrifuged at 15 000× g at 4 °C for 5 min. The supernatants were added to 50 mg of Ral GDS-RBD agarose beads (glutathione-sepharose beads pre-coupled to GST fused to the Ras-binding domain of Ral GDS) and incubated at 4 °C for 45 min with gentle rotation. The beads were then washed four times in lysis buffer and boiled in SDS sample buffer. The amount of GTP-bound Rap1 was analyzed by immunoblotting with an anti-Rap1 antibody (Santa Cruz).
Abbreviations
- BSA:
-
(bovine serum albumin)
- DMEM:
-
(Dulbecco's modified Eagle's medium)
- GDNF:
-
(glial cell line-derived neurotrophic factor)
- EGFP:
-
(enhanced green fluorescent protein)
- FBS:
-
(fetal bovine serum)
- PBS:
-
(phosphate-buffered saline)
- RTK:
-
(receptor tyrosine kinase)
- SDS-PAGE:
-
(sodium dodecyl sulfate polyacrylamide gel electrophoresis)
References
Lin LF, Doherty DH, Lile JD, Bektesh S, Collins F . GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science 1993; 260:1130–1132.
Airaksinen MS, Saarma M . The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci 2002; 3:383–394.
Sariola H, Saarma M . Novel functions and signalling pathways for GDNF. J Cell Sci 2003; 116:3855–3862.
Runeberg-Roos P, Saarma M . Neurotrophic factor receptor RET: structure, cell biology, and inherited diseases. Ann Med 2007; 39:572–580.
Sariola H, Sainio K . The tip-top branching ureter. Curr Opin Cell Biol 1997; 9:877–884.
Meng X, Lindahl M, Hyvönen ME, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 2000; 287:1489–1493.
Deierborg T, Soulet D, Roybon L, Hall V, Brundin P . Emerging restorative treatments for Parkinson's disease. Prog Neurobiol 2008; 85:407–432.
Hong M, Mukhida K, Mendez I . GDNF therapy for Parkinson's disease. Expert Rev Neurother 2008; 8:1125–1139.
Jing S, Wen D, Yu Y, et al. GDNF-induced activation of the ret protein tyrosine kinase is mediated by GDNFR-alpha, a novel receptor for GDNF. Cell 1996; 85:1113–1124.
Paratcha G, Ledda F, Ibáñez CF . The neural cell adhesion molecule NCAM is an alternative signaling receptor for GDNF family ligands. Cell 2003; 113:867–879.
Schuchardt A, D'Agati V, Larsson-Blomberg L, Costantini F, Pachnis V . Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 1994; 367:380–383.
Kramer ER, Aron L, Ramakers GM, et al. Absence of Ret signaling in mice causes progressive and late degeneration of the nigrostriatal system. PLoS Biol 2007; 5:e39.
Edery P, Lyonnet S, Mulligan LM, et al. Mutations of the RET proto-oncogene in Hirschsprung's disease. Nature 1994; 367:378–380.
Romeo G, Ronchetto P, Luo Y, et al. Point mutations affecting the tyrosine kinase domain of the RET proto-oncogene in Hirschsprung's disease. Nature 1994; 367:377–378.
Lai AZ, Gujral TS, Mulligan LM . RET signaling in endocrine tumors: delving deeper into molecular mechanisms. Endocr Pathol 2007; 18:57–67.
Besset V, Scott RP, Ibáñez CF . Signaling complexes and protein-protein interactions involved in the activation of the Ras and phosphatidylinositol 3-kinase pathways by the c-Ret receptor tyrosine kinase. J Biol Chem 2000; 275:39159–39166.
Hayashi Y, Iwashita T, Murakamai H, et al. Activation of BMK1 via tyrosine 1062 in RET by GDNF and MEN2A mutation. Biochem Biophys Res Commun 2001; 281:682–689.
Takahashi M . The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev 2001; 12:361–373.
Manié S, Santoro M, Fusco A, Billaud M . The RET receptor: function in development and dysfunction in congenital malformation. Trends Genet 2001; 17:580–589.
Fukuda T, Kiuchi K, Takahashi M . Novel mechanism of regulation of Rac activity and lamellipodia formation by RET tyrosine kinase. J Biol Chem 2002; 277:19114–19121.
Uchida M, Enomoto A, Fukuda T, et al. Dok-4 regulates GDNF-dependent neurite outgrowth through downstream activation of Rap1 and mitogen-activated protein kinase. J Cell Sci 2006; 119:3067–3077.
Zhang Y, Zhu W, Wang YG, et al. Interaction of SH2-Bbeta with RET is involved in signaling of GDNF-induced neurite outgrowth. J Cell Sci 2006; 119:1666–1676.
Rubinfe B, Wong G, Bekesi E, et al. A synthetic peptide corresponding to a sequence in the GTPase activating protein inhibits p21 ras stimulation and promotes guanine nucleotide exchange. Int J Pept Protein Res 1991; 38:47–53.
Bos JL, de Rooij J, Reedquist KA . Rap1 signalling: adhering to new models. Nat Rev Mol Cell Biol 2001; 2:369–377.
Kometani K, Ishida D, Hattori M, Minato N . Rap1 and SPA-1 in hematologic malignancy. Trends Mol Med 2004; 10:401–408.
Gao Q, Kumar A, Singh L, et al. Human papillomavirus E6-induced degradation of E6TP1 is mediated by E6AP ubiquitin ligase. Cancer Res 2002; 62:3315–3321.
Chen Y, Wang PY, Ghosh A . Regulation of cortical dendrite development by Rap1 signaling. Mol Cell Neurosci 2005; 28:215–228.
Trupp M, Belluardo N, Funakoshi H, Ibáñez CF . Complementary and overlapping expression of glial cell line-derived neurotrophic factor (GDNF), c-ret proto-oncogene, and GDNF receptor-alpha indicates multiple mechanisms of trophic actions in the adult rat CNS. J Neurosci 1997; 17:3554–3567.
Kodama Y, Asai N, Kawai K, et al. The RET proto-oncogene: a molecular therapeutic target in thyroid cancer. Cancer Sci 2005; 96:143–148.
Hanks SK, Quinn AM, Hunter T . The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 1988; 241:42–52.
Kaplan DR, Miller FD . Neurotrophin signal transduction in the nervous system. Curr Opin Neurobiol 2000; 10:381–391.
Vaudry D, Chen Y, Hsu CM, Eiden LE . PC12 cells as a model to study the neurotrophic activities of PACAP. Ann N Y Acad Sci 2002; 971:491–496.
Bos JL, de Bruyn K, Enserink J, et al. The role of Rap1 in integrin-mediated cell adhesion. Biochem Soc Trans 2003; 31 (Pt 1):83–86.
Jordan JD, Carey KD, Stork PJ, Iyengar R . Modulation of rap activity by direct interaction of Galpha(o) with Rap1 GTPase-activating protein. J Biol Chem 1999; 274:21507–21510.
Mochizuki N, Ohba Y, Kiyokawa E, et al. Activation of the ERK/MAPK pathway by an isoform of rap1GAP associated with G alpha(i). Nature 1999; 400:891–894.
Jordan JD, He JC, Eungdamrong NJ, et al. Cannabinoid receptor-induced neurite outgrowth is mediated by Rap1 activation through G(alpha)o/i-triggered proteasomal degradation of Rap1GAPII. J Biol Chem 2005; 280:11413–11421.
Gérard A, Favre C, Garcon F, et al. Functional interaction of RasGAP-binding proteins Dok-1 and Dok-2 with the Tec protein tyrosine kinase. Oncogene 2004; 23:1594–1598.
Jones N, Dumont DJ . Recruitment of Dok-R to the EGF receptor through its PTB domain is required for attenuation of Erk MAP kinase activation. Curr Biol 1999; 9:1057–1060.
Meakin SO, MacDonald JI, Gryz EA, Kubu CJ, Verdi JM . The signaling adapter FRS-2 competes with Shc for binding to the nerve growth factor receptor TrkA. A model for discriminating proliferation and differentiation. J Biol Chem 1999; 274:9861–9870.
Sasagawa S, Ozaki Y, Fujita K, Kuroda S . Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat Cell Biol 2005; 7:365–373.
Trupp M, Rydén M, Jörnvall H, et al. Peripheral expression and biological activities of GDNF, a new neurotrophic factor for avian and mammalian peripheral neurons. J Cell Biol 1995; 130:137–148.
Pong K, Xu RY, Beck KD, Zhang TJ, Louis JC . Inhibition of glial cell line-derived neurotrophic factor induced intracellular activity by K-252b on dopaminergic neurons. J Neurochem 1997; 69:986–994.
Zurn AD, Winkel L, Menoud A, Djabali K, Aebischer P . Combined effects of GDNF, BDNF, and CNTF on motoneuron differentiation in vitro. J Neurosci Res 1996; 44:133–141.
Gavazzi I, Kumar RD, McMahon SB, Cohen J . Growth responses of different subpopulations of adult sensory neurons to neurotrophic factors in vitro. Eur J Neurosci 1999; 11:3405–3414.
Schäfer KH, Mestres P . The GDNF-induced neurite outgrowth and neuronal survival in dissociated myenteric plexus cultures of the rat small intestine decreases postnatally. Exp Brain Res 1999; 125:447–452.
Kirik D, Georgievska B, Björklund A . Localized striatal delivery of GDNF as a treatment for Parkinson disease. Nat Neurosci 2004; 7:105–110.
Cao L, Liu L, Chen ZY, et al. Olfactory ensheathing cells genetically modified to secrete GDNF to promote spinal cord repair. Brain 2004; 127:535–549.
Love S, Plaha P, Patel NK, Hotton GR, Brooks DJ, Gill SS . Glial cell line-derived neurotrophic factor induces neuronal sprouting in human brain. Nat Med 2005; 11:703–704.
Markus A, Patel TD, Snider WD . Neurotrophic factors and axonal growth. Curr Opin Neurobiol 2002; 12:523–531.
Segal RA . Selectivity in neurotrophin signaling: theme and variations. Annu Rev Neurosci 2003; 26:299–330.
Zhang Y, Yan Z, Farooq A, et al. Molecular basis of distinct interactions between Dok1 PTB domain and tyrosine phosphorylated EGF receptor. J Mol Biol 2004; 343:1147–1155.
Mise N, Drosten M, Racek T, Tannapfel A, Pützer BM . Evaluation of potential mechanisms underlying genotype-phenotype correlations in multiple endocrine neoplasia type 2. Oncogene 2006; 25:6637–6647.
Mologni L, Sala E, Cazzaniga S, et al. Inhibition of RET tyrosine kinase by SU5416. J Mol Endocrinol 2006; 7:199–212.
Kramer ER, Aron L, Ramakers GM, et al. Absence of Ret signaling in mice causes progressive and late degeneration of the nigrostriatal system. PLoS Biol 2007; 5:e39.
Miyata M, Rikitake Y, Takahashi M, et al. Regulation by afadin of cyclical activation and inactivation of Rap1, Rac1, and RhoA small G proteins at leading edges of moving NIH3T3 cells. J Biol Chem 2009; 284:24595–24609.
Acknowledgements
We are grateful to Dr Carlos F Ibanez (Karolinska Institute, Sweden) for generously providing the RET cDNA. The Rap1GAP cDNA was a generous gift from Dr Patrick J Casey (Duke University Medical Center, USA). The GST-RalGDS-RBD plasmid (for detecting Rap1 activity) was a generous gift from Dr Chengbiao Wu (Stanford University School of Medicine, USA). This work was supported by the National Natural Science Foundation of China (30400123, 30570939, 30770657 and 30530240); National Key Basic Research Program (2006CB500702, 2007CB947100, 2011CB504401 and 2009ZX09311-001); Shanghai Rising-Star Program (05QMX1469) and Shanghai Metropolitan Fund for Research and Development (07DJ14005).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jiao, L., Zhang, Y., Hu, C. et al. Rap1GAP interacts with RET and suppresses GDNF-induced neurite outgrowth. Cell Res 21, 327–337 (2011). https://doi.org/10.1038/cr.2010.139
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2010.139
Keywords
This article is cited by
-
Intracellular RET signaling pathways activated by GDNF
Cell and Tissue Research (2020)
-
The multi-targeted tyrosine kinase inhibitor vandetanib plays a bifunctional role in non-small cell lung cancer cells
Scientific Reports (2015)
-
Neural protein gamma-synuclein interacting with androgen receptor promotes human prostate cancer progression
BMC Cancer (2012)
-
Neuroscience in China 2010–2011
Chinese Science Bulletin (2012)
-
Positive autoregulation of GDNF levels in the ventral tegmental area mediates long-lasting inhibition of excessive alcohol consumption
Translational Psychiatry (2011)