ABSTRACT
IAPs (inhibitors of apoptosis) are a family of proteins containing one or more characteristic BIR domains. These proteins have multiple biological activities that include binding and inhibiting caspases, regulating cell cycle progression, and modulating receptor-mediated signal transduction. Our recent studies found the IAP family members XIAP and c-IAP1 are ubiquitinated and degraded in proteasomes in response to apoptotic stimuli in T cells, and their degradation appears to be important for T cells to commit to death. In addition to three BIR domains, each of these IAPs also contains a RING finger domain. We found this region confers ubiquitin protease ligase (E3) activity to IAPs, and is responsible for the auto-ubiquitination and degradation of IAPs after an apoptotic stimulus. Given the fact that IAPs can bind a variety of proteins, such as caspases and TRAFs, it will be of interest to characterize potential substrates of the E3 activity of IAPs and the effects of ubiquitination by IAPs on signal transduction, cell cycle, and apoptosis.
Similar content being viewed by others
INTRODUCTION
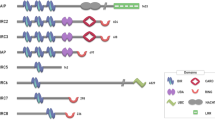
The iap (inhibitor of apoptosis) gene was first identified in baculovirus because it protected infected cells from death and enhanced virus replication1. IAPs are now known to be expressed in organisms ranging from yeast to mammals2,3. All the IAPs contain one to three BIR (baculovirus IAP repeats) domains, each consisting of approximately 70 amino acid residues. Many IAPs also have a RING finger domain, defined by seven cysteines and one histidine (e.g. C3HC4) that can coordinate two zinc atoms4. Among the mammalian IAPs, c-IAP1, cIAP2, and XIAP have three BIRs in the N-terminal portion of the molecule and a RING finger at the C-terminus, NAIP contains three BIRs without RING, and survivin and BRUCE each has just one BIR.
The structure of the third BIR of c-IAP1 has been determined by nuclear magnetic resonance5. The 70-residue region forms four short α-helices linked by a number of loops. In the core of this BIR there are extensive interactions between the large number of hydrophobic residues and a zinc atom coordinated by three cysteines and a histidine. On the surface of the BIR domain, a number of hydrophobic regions and conserved charged amino acids have been identified, which may participate in interactions of IAPs with other proteins. Like the c-IAP1 BIR domain, the NMR structure of the second BIR of XIAP contains four a-helices, a hydrophobic zinc-containing core, and a number of conserved charged residues on its surface6. The major difference is that the BIR of XIAP has three anti-parallel b-sheets that are not found in the BIR of c-IAP1. Whether this is related to the functional differences between different BIRs remains to be determined.
IAPs bind and inhibit caspases
XIAP, c-IAP1, and c-IAP2 can directly bind to activated caspase-3 and -7 and inhibit their activities7,8 Mutational analysis indicated that the BIR domains are responsible for the inhibition2,3. Further experiments with XIAP found the second BIR, but not the first or the third BIR, is able to bind and inhibit caspase-3 and-7 with an apparent inhibition constant of 2-5 n M9. XIAP, c-IAP1, and c-IAP2 can also inhibit activated caspase-9, and in addition, they bind pro-caspase 9 and prevent its activation by apoptotic stimuli10. Interestingly, a BIR3-RING fragment, but not a BIR1-BIR2 fragment of XIAP, binds and inhibits caspase-911, indicating the BIR domains of XIAP selectively interact with various caspases. Consistent with these results, lepidopteran IAP (sfIAP) and baculoviral IAP (CpIAP), both of which have two BIRs and a RING domain, are inhibitors of mammalian caspase-9 but not caspase-3 or -712. The mechanisms of the specificity of various BIRs for caspases are not well understood at this time. It has been shown that several residues in the linker region N-terminal to the BIR2 domain of XIAP are critical for inhibiting caspase-3, even though they do not affect the structure of the BIR6. A chimeric molecule containing this linker region and XIAP BIR1 is able to inhibit caspase-3, although the linker region or BIR1 alone can not inhibit the caspase6. Further experiments indicated that the XIAP BIR2 domain interacts with both pro-caspase-3 and activated caspase-3, whereas the linker region only interacts with activated caspase6. Since caspase-3 acts on consensus sequence DEVD, it has been suggested that the DxxD sequence in this linker region directly interacts with the active center of caspase-3 and blocks its activity6. It will be of interest to determine whether the inhibition of caspase-9 by XIAP and inhibition of caspaspe-3, -7, and -9 by c-IAP1 and c-IAP2 also depend on residues around the corresponding BIRs. This might also help to explain why BIR1- BIR2 and BIR3- RING of XIAP interact specifically with caspase-3 and caspase-9 respectively.
IAPs and signaling of membrane receptors
The RING-containing c-IAP1 and c-IAP2 were first identified as components of the type II TNF receptor (TNFR ) signaling complex. They were recruited to the TNFR by interactions between their BIRs and TRAF (TNFR-associated factor)-1 and -2, which directly bind to the TNFR13. In sensitive cells, TNF treatment leads to apoptosis, which is mediated by activation of caspase-8 mediated by interactions between the death domains of TNFR and adapter molecules (TRADD and FADD)14. Although c-IAP1 and c-IAP2 have been shown to inhibit TNF-induced apoptosis in transient transfection systems15,16, it is not known whether this is due to their caspase-inhibitory activity, their involvement in TNFR-mediated signaling, or both. Another prominent activity of TNF signaling is induction of transcription factor NFkB, which suppresses apoptosis17. TRAF1, TRAF2, c-IAP1, and c-IAP2 all are target genes of NFκB16, and expression of all these molecules is required to effectively block TNF-induced cell death in NFκB deficient cells. It has also been shown that activation of NFκB is required for its inhibition of TNF-induced cell death by over-expression of c-IAP215. Thus, IAPs could be induced by NFκB, and may in turn signal through this transcription factor to inhibit apoptosis. TNF also activates the transcription factor AP-1, which plays an important role in mediating the effects of TNF on the expression of other genes and cytokines14. TNF induction of AP-1 is largely due to activation of c-Jun N-terminal kinase (JNK)18. Genetic experiments demonstrated that TRAF2 is required for TNFR to activate JNK19. Although it also contains a RING domain, TRAF2 can not catalyze its own ubiquitination (Li et al, unpublished observation). Given its association with cIAP1 and cIAP2, it is interesting to know whether cIAPs can ubiquitinate TRAF2 and regulate its intracellular level.
Other IAPs are also involved in receptor-mediated signaling processes. For example, the Drosophila IAPs D-IAP1 and D-IAP2 interact with the receptor for decapentaplegic, a Drosophila bone morphogenetic protein (BMP)/TGF-β superfamily member20. This interaction appears to be mediated through the RING domain of D-IAPs, but its functional consequence has not been determined. In a different system, XIAP binds directly to BMP receptor and its downstream signaling molecule TAB1, an activator of a MAP kinase kinase kinase family member named TAK121. The activation of TAB1-TAK1 promotes ventral mesoderm formation in early Xenopus embryos, which is enhanced by overexpression of wild type XIAP and blocked by expression of an N-terminal truncated XIAP21. Therefore, XIAP may function as an adapter protein for BMP receptor signaling. Conversely, transfection of XIAP can selectively activate the JNK1 kinase22, which is also activated upon stimulation of BMP/TGF-β receptor23. It has been shown that the BIR domain of XIAP is responsible for association with TAB1 and activation JNK in these two systems21,22. Interestingly, a dominant negative form of JNK1 has been shown to suppress the anti-apoptotic action of XIAP in caspase-1-induced apoptosis22, suggesting JNK1 mediates the anti-apoptotic action of XIAP in this system.
IAPs and cell cycle control
Survivin has only one BIR and is abundantly expressed during fetal development, but rarely present in adult tissues24. It is also expressed in most transformed cell lines, but not in non-transformed human peripheral T cells, fibroblasts, or endothelial cells. Overexpression of survivin in an IL-3-dependent pre-B cell line significantly inhibited cell death induced by withdrawal of IL-324. Further analysis indicated that survivin can partially block activation of caspases and cell death induced by a variety of stimuli, such as Bax, Fas engagement, and etoposide25. This appears to be due to its ability to bind to activated caspases25,26. Survivin expression is also closely connected with cell cycle. Studies using HeLa cells found that the expression of survivin is upregulated ∼ 40-fold in G2/M27. At the beginning of mitosis, survivin associates specifically with microtubules of the mitotic spindle. Mutated survivin that can not bind microtubules lost its anti-apoptotic activity27. Furthermore, inhibition of survivin expression by antisense oligonucleotides or transfection of dominant negative survivin induced apoptosis and formation of multinucleated polyploid cells28,29. This is presumably due to failed cytokinesis, the process of separating a cell into two after it has duplicated its genetic material. It has also been shown that the murine homolog of survivin is highly expressed in thymocytes and induced in peripheral T cells upon activation26, indicating that survivin is also associated with cell cycle progression in untransformed cells. In addition, C. elegans embryos that lack the survivin homologue bir-1 were unable to complete cytokinesis and became multinucleated30. This cytokinesis defect could be partially suppressed by transgenic expression of survivin. In the yeast S. cerevisiae and S. probe there is only a single IAP protein, Bir1, which contains two BIRs31. This gene is required for spindle elongation and efficient meiotic division31, indicating the cell cycle regulation function of IAP family proteins is evolutionarily conserved. It is not clear at present whether the cell-cycle regulating activity of these BIR-containing proteins is related to their anti-apoptotic function.
IAPs and protein ubiquitination
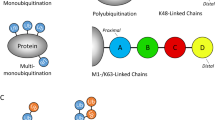
Protein degradation is essential for many cellular functions, including cell cycle control, removal of misfolded or damaged proteins, activation of transcription factors, and generation of antigen peptides for MHC molecules to present to T cells32,33. The multisubunit protease complex, known as the proteasome, is one of the major sites in eukaryotic cells where these tasks are accomplished34. Proteasomes recognize and degrade proteins that have been covalently modified with polymerized chains of ubiquitin, an 8 kDa protein that is ubiquitously expressed and evolutionarily conserved. Ubiquitin is activated to a high-energy thioester at its C-terminal Gly residue by ubiquitin-activating enzyme (E1). In concert with ubiquitin-conjugating enzyme (E2 or UBC), a ubiquitin protein ligase (E3) transfers the activated ubiquitin to a Lys residue in the substrate that has been recognized and bound by the E3. Polyubiquitin chains are synthesized by adding subsequent activated ubiquitins to the Lys residue of the previously conjugated ubiquitin34. In this enzyme system, a single E1 can carry out the activation of ubiquitin, required for all subsequent reactions. Over 14 different E2 molecules have been identified, and each E2 enzyme can act with more than one type of E3. However, there may be many more E3 enzymes in cells because each E3 has to recognize specific substrates. Thus, identification and characterization of E3s is critical in understanding the ubiquitination and degradation of specific proteins.
Protein degradation also plays an important role in T cell apoptosis35. This is exemplified by the finding that proteasome inhibitors can effectively block thymocyte apoptosis induced by various apoptotic stimuli36. It is conceivable that this effect of proteasome inhibition is due to the inhibition of degradation of proteins that prevent cell death. Studies based on this premise have recently demonstrated that XIAP, c-IAP1, and c-IAP2 are degraded in a proteasome-dependent manner when thymocytes are induced to undergo apoptosis by glucocorticoids or etoposide37. Furthermore, these IAPs have E3 activity and are able to catalyze their own ubiquitination in vitro and in transfected cells. This E3 activity depends on the RING domain, as the change of a single zinc-chelating residue in this region can eliminate the E3 activity37. T cell hybridomas transfected with a truncated XIAP that lacks the RING domain are more resistant to glucocorticoid-induced apoptosis than cells transfected with wild type XIAP37. Consistent with these results, it has been shown that remove of the RING domain of Drosophila IAPs (D-IAP1 and D-IAP2) enhanced their ability to inhibit cell death in eye caused by expression of cell death-inducing protein Reaper38. These results indicate that autoubiquitination of IAPs may play an important role in their own degradation, allowing cells to commit to apoptosis.
BRUCE (BIR repeat containing ubiquitin-conjugating enzyme) is a giant protein (530 kDa) associated with the membrane of the Golgi system39. The human homolog of this protein is named Apollon40. BRUCE and Apollon have one BIR domain, but it has not yet known if these molecules can bind caspases or inhibit apoptosis. However, cells treated with Apollon antisense oligonucleotides had reduced the expression of the protein, and were sensitized to undergo apoptosis induced by chemotherapeutic agents40, suggesting that BRUCE and Apollon may function as inhibitors of apoptosis. BRUCE also contains at its C-terminus a typical UBC domain, an E2 hallmark that harbors the active site cysteine required for thioester formation41. BRUCE was found to form a conjugate with ubiquitin in the presence of E1 in vitro, confirming its E2 activity39. It is not known yet if BRUCE functions with an E3, what proteins it might ubiquitinate, and what its physiological function might be.
It is worth noting that more than 200 proteins with many different functions have a RING finger domain4. Several well-known multi-subunits E3 contain RING proteins that are required for their activities42. It appears that the role of RING proteins in these E3s is related to the assembly of these complexes, but not directly involved in substrate recognition or ubiquitin transfer4. However, like the IAP proteins, a number of RING-containing protein themselves possess E3 activity that depends upon an intact RING domain43, 44, 45. For example, the RING-containing protein Cbl mediates the ubiquitination of the receptor for epidermal growth factor (EGFR)43,44. The product of the breast cancer susceptibility gene brac1 promotes autoubiquitination43. The p53-binding protein Mdm2 can catalyze both autoubiquitination and ubiquitination of its substrate, p5345. These results suggest that the RING domain may directly participate the transfer of ubiquitin from E2 to substrate. Furthermore, ubiquitination of EGFR and p53 by Cbl and Mdm2, respectively, modulates their intracellular levels45, 46, 47, indicating that these ubiquitination reactions are specific and physiologically important. This is strongly supported by the finding that the E3 activity of IAPs is important in regulating their own level and apoptosis-inhibiting activity37. In addition, it is conceivable that XIAP, c-IAP1, and c-IAP2 may not only regulate their own levels in cells, but may also modulate the amount of their (putative) substrates. Given the fact that IAPs associate with many proteins, such as caspases, TRAF-2, and TGF-β receptor, it will be interesting to determine if these proteins are ubiquitinated by IAPs, and if so what effect this might have on signal transduction and cell fate.
Conclusion
The IAPs family proteins are known for their ability to inhibit apoptosis induced through mitochondria-dependent and-independent pathways. It appears that this can be attributed to their ability to prevent activation of procaspase-9, and inhibit caspase-3, -7, and -9. However, there is also compelling evidence indicating that the inhibition of c-IAP1 and c-IAP2 on TNF-induced apoptosis is related to their affect on TNFR-mediated induction of NFkB. Both the mammalian IAP survivin and the yeast IAP Bir-1 have profound effects on cell cycle progression. This might be the major function of this type of IAP, as yeast does not express caspases. We have recently found that RING finger-containing IAPs have E3 activity and can catalyze their own ubiquitination. This activity appears to be important in regulating the intracellular level of these IAPs and their effects on apoptosis. These results not only demonstrate that the RING finger domain participates in protein ubiquitin ligation, but also raise the possibility that the E3 activity of these IAPs may be involved in the multiple biological activities of these IAPs in cells. For example, it has been shown recently that cIAP1 is able to catalyze in vitro ubiquitination of caspase-3 and -7, but not caspase-148. Although it remains to be determined whether this ubiquitination can happen in cells, the results raise the possibility that the E3 activity of cIAP1 may contribute to its inhibition on caspases.
Abbreviations
- IAP:
-
inhibitors of apoptosis
- BIR:
-
baculovirus IAP repeats
- E1:
-
ubiquitin-activating enzyme
- E2:
-
ubiquitin-conjugating enzyme
- E3:
-
ubiquitin protein ligase
- TNFR:
-
tumor necrosis factor receptor
- TRAF2:
-
tumor necrosis factor receptor associated factor-2
- JNK:
-
c-Jun N-terminal kinase
References
Crook NE, Clem RJ, Miller LK . An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol 1993; 67:2168–74.
Deveraux QL, Reed JC . IAP family proteins-suppressors of apoptosis. Genes Dev. 1999; 13:239–52.
Miller LK . An exegesis of IAPs: salvation and surprises from BIR motifs. Trends Cell Biol 1999; 9:323–8.
Borden KL . RING domains: master builders of molecular scaffolds? J Mol Biol 2000; 295:1103–12.
Hinds MG, Norton RS, Vaux DL, Day CL . Solution structure of a baculoviral inhibitor of apoptosis (IAP) repeat. Nat Struct Biol 1999; 6:648–51.
Sun C, Cai M, Gunasekera AH, et al. NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature 1999; 401:818–22.
Deveraux QL, Takahashi R, Salvesen GS, Reed JC . X-linked IAP is a direct inhibitor of cell-death proteases. Nature 1997; 388:300–4.
Roy N, Deveraux QL, Takahashi R, Salvesen GS, Reed JC . The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J 1997; 16:6914–25.
Takahashi R, Deveraux Q, Tamm I, et al. A single BIR domain of XIAP sufficient for inhibiting caspases. J Biol Chem 1998; 273:7787–90.
Deveraux QL, Roy N, Stennicke HR, et al. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J 1998; 17:2215–23.
Deveraux QL, Leo E, Stennicke HR, Welsh K, Salvesen GS, Reed JC . Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. EMBO J 1999; 18:5242–51.
Huang Q, Deveraux QL, Maeda S, et al. Evolutionary conservation of apoptosis mechanisms: lepidopteran and baculoviral inhibitor of apoptosis proteins are inhibitors of mammalian caspase-9. Proc Natl Acad Sci USA 2000; 97:1427–32.
Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV . The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell 1995; 83:1243–52.
Kaufman DR, Choi Y . Signaling by tumor necrosis factor receptors: pathways, paradigms and targets for therapeutic modulation. Int Rev Immunol 1999; 18:405–27.
Chu ZL, McKinsey TA, Liu L, Gentry JJ, Malim MH, Ballard DW . Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-kappaB control. Proc Natl Acad Sci USA 1997; 94:10057–62.
Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin ASJ . NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 1998; 281:1680–3.
Van Antwerp DJ, Martin SJ, Verma IM, Green DR . Inhibition of TNF-induced apoptosis by NF-kappa B. Trends Cell Biol 1998; 8:107–11.
Westwick JK, Weitzel C, Minden A, Karin M, Brenner DA . Tumor necrosis factor alpha stimulates AP-1 activity through prolonged activation of the c-Jun kinase. J Biol Chem 1994; 269:26396–401.
Yeh WC, Shahinian A, Speiser D, et al. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity 1997; 7:715–25.
Oeda E, Oka Y, Miyazono K, Kawabata M . Interaction of Drosophila inhibitors of apoptosis with thick veins, a type I serine/threonine kinase receptor for decapentaplegic. J Biol Chem 1998; 273:9353–6.
Yamaguchi K, Nagai S, Ninomiya-Tsuji J, et al. XIAP, a cellular member of the inhibitor of apoptosis protein family, links the receptors to TAB1-TAK1 in the BMP signaling pathway. EMBO J 1999; 18:179–87.
Sanna MG, Duckett CS, Richter BW, Thompson CB, Ulevitch RJ . Selective activation of JNK1 is necessary for the anti-apoptotic activity of hILP. Proc Natl Acad Sci USA 1998;,b> 95:6015–20.
Massague J . TGF-beta signal transduction. Annu Rev Biochem 1998; 67:753–91.
Ambrosini G, Adida C, Altieri DC . A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 1997; 3:917–21.
Tamm I, Wang Y, Sausville E, et al. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res 1998; 58:5315–20.
Kobayashi K, Hatano M, Otaki M, Ogasawara T, Tokuhisa T . Expression of a murine homologue of the inhibitor of apoptosis protein is related to cell proliferation. Proc Natl Acad Sci USA 1999; 96:1457–62.
Li F, Ambrosini G, Chu EY, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 1998; 396:580–4.
Ambrosini G, Adida C, Sirugo G, Altieri DC . Induction of apoptosis and inhibition of cell proliferation by survivin gene targeting. J Biol Chem 1998; 273:11177–82.
Li F, Ackermann EJ, Bennett CF, et al. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat Cell Biol 1999; 1:461–6.
Fraser AG, James C, Evan GI, Hengartner MO . Caenorhabditis elegans inhibitor of apoptosis protein (IAP) homologue BIR-1 plays a conserved role in cytokinesis. Curr Biol 1999; 9:292–301.
Uren AG, Beilharz T, O'Connell MJ, et al. Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division. Proc Natl Acad Sci USA 1999; 96:10170–75.
Weissman AM . Regulating protein degradation by ubiquitination. Immunol Today 1997; 18:189–98.
Rock KL, Goldberg AL . Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol 1999; 17:739–79.
Hershko A, Ciechanover A . The ubiquitin system. Annu Rev Biochem 1998; 67:425–79.
Orlowski RZ . The role of the ubiquitin-proteasome pathway in apoptosis. Cell Death Differ 1999; 6:303–13.
Grimm LM, Goldberg AL, Poirier GG, Schwartz LM, Osborne BA . Proteasomes play an essential role in thymocyte apoptosis. EMBO J 1996; 15:3835–44.
Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD . Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science 2000; 288:874–7.
Hay BA, Wassarman DA, Rubin GM . Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 1995; 83:1253–62.
Hauser HP, Bardroff M, Pyrowolakis G, Jentsch S . A giant ubiquitin-conjugating enzyme related to IAP apoptosis inhibitors. J Cell Biol 1998; 141:1415–22.
Chen Z, Naito M, Hori S, Mashima T, Yamori T, Tsuruo T . A human IAP-family gene, apollon, expressed in human brain cancer cells. Biochem Biophys Res Commun 1999; 264:847–54.
Jentsch S, Seufert W, Sommer T, Reins HA . Ubiquitin- conjugating enzymes: novel regulators of eukaryotic cells. Trends Biochem Sci 1990; 15:195–98.
Tyers M, Willems AR . One ring to rule a superfamily of E3 ubiquitin ligases. Science 1999; 284:601–4.
Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM . RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA 1999; 96:11364–69.
Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC . The tyrosine kinase negative regulator c-Cbl as a RING-type, E2- dependent ubiquitin- protein ligase [see comments]. Science 1999; 286:309–12.
Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM . Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 2000; 275:8945–51.
Waterman H, Levkowitz G, Alroy I, Yarden Y . The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J Biol Chem 1999; 274:22151–54.
Yokouchi M, Kondo T, Houghton A, et al. Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J Biol Chem 1999; 274:31707–12.
HK H, Joazeiro CA, Bonfoco E, Kamada S, Leverson JD, Hunter T . The Inhibitor of Apoptosis, cIAP1, Functions as a Ubiquitin-Protein Ligase and Promotes in vitro Ubiquitination of caspases-3 and -7. J Biol Chem 2000.
Acknowledgements
We thank Dr. Jonathan D. Ashwell for discussions and critical reading of this review.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
YANG, Y., LI, X. The IAP family: endogenous caspase inhibitors with multiple biological activities. Cell Res 10, 169–177 (2000). https://doi.org/10.1038/sj.cr.7290046
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7290046
Keywords
This article is cited by
-
BRD7 inhibits enhancer activity and expression of BIRC2 to suppress tumor growth and metastasis in nasopharyngeal carcinoma
Cell Death & Disease (2023)
-
The Emerging Roles of E3 Ligases and DUBs in Neurodegenerative Diseases
Molecular Neurobiology (2023)
-
Increased migration and motility in XIAP-null cells mediated by the C-RAF protein kinase
Scientific Reports (2022)
-
DCAF12 promotes apoptosis and inhibits NF-κB activation by acting as an endogenous antagonist of IAPs
Oncogene (2022)
-
RNA-Seq analysis reveals potential regulators of programmed cell death and leaf remodelling in lace plant (Aponogeton madagascariensis)
BMC Plant Biology (2021)