Abstract
Caspases are the executioners of apoptosis. Although much is known about their physiological roles and structures, detailed analyses of missense mutations of caspases are lacking. As mutations within caspases are identified in various human diseases, the study of caspase mutants will help to elucidate how caspases interact with other components of the apoptosis pathway and how they may contribute to disease. DrICE is the major effector caspase in Drosophila required for developmental and stress-induced cell death. Here, we report the isolation and characterization of six de novo drICE mutants, all of which carry point mutations affecting amino acids conserved among caspases in various species. These six mutants behave as recessive loss-of-function mutants in a homozygous condition. Surprisingly, however, two of the newly isolated drICE alleles are gain-of-function mutants in a heterozygous condition, although they are loss-of-function mutants homozygously. Interestingly, they only behave as gain-of-function mutants in the presence of an apoptotic signal. These two alleles carry missense mutations affecting conserved amino acids in close proximity to the catalytic cysteine residue. This is the first time that viable gain-of-function alleles of caspases are described in any intact organism and provides a significant exception to the expectation that mutations of conserved amino acids always abolish the pro-apoptotic activity of caspases. We discuss models about how these mutations cause the gain-of-function character of these alleles.
Similar content being viewed by others
Main
Apoptosis is a major form of programmed cell death.1 The core apoptotic machinery is evolutionary conserved with caspases as the fundamental components.1, 2, 3 Caspases are specific cysteine proteases that are produced as inactive zymogens composed of an N-terminal pro-domain, a large subunit region with the catalytic cysteine residue, and a small subunit region at the carboxyl end.2, 4 Depending on their structures and functions, caspases are grouped into initiator and effector caspases.2, 3 Initiator caspases possess long pro-domains, which facilitate the recruitment of initiator caspases into cell death signaling complexes for activation.2, 3, 5 Effector caspases are activated by initiator caspase complexes through proteolytic processing, cleaving off the pro-domain and separating the large and small subunits. The active effector caspase is a tetramer composed of two large and two small subunits and contains two catalytic sites.2, 3 Activated effector caspases cleave many protein targets to trigger the physiological and morphological changes characteristic of apoptosis. In mammals, apoptotic initiator caspases are Caspase-8, -9, -2 and -10, and effector caspases involved in apoptosis are Caspase-3, -7 and -6.2, 3 Of the seven Drosophila caspases, only the initiator caspase Dronc (Caspase-9-like) and the effector caspases DrICE and Dcp-1 (Caspase-3-like) have been implicated in apoptosis in imaginal discs.1, 2, 6
Caspase activation is tightly regulated in surviving cells. Inhibitor of apoptosis proteins (IAPs) directly bind to and inhibit processed caspases.7, 8, 9 The best-characterized IAPs are mammalian XIAP and Drosophila IAP1 (DIAP1).10, 11 In cells committed to apoptosis, IAP-mediated inhibition of caspases is counteracted by IAP antagonists. Specifically, the IAP antagonists encoded by reaper, hid and grim (RHG)12, 13, 14, 15 in Drosophila trigger proteolytic degradation of DIAP116, 17, 18, 19, 20 which releases caspases from DIAP1 inhibition and triggers apoptosis. The overexpression of the RHG genes in the fly eye using the eye-specific GMR promoter causes an eye ablation phenotype due to massive apoptosis (see, for example, GMR-hid in Figure 1a).12, 13, 14 In fact, mutants of diap1, dronc and drICE genes were isolated in genetic screens searching for modifiers of the eye ablation phenotypes caused by reaper or hid overexpression.21, 22, 23, 24, 25, 26, 27 Mammalian IAP antagonists are Smac/Diablo and HtrA2/Omi, which function similarly to the RHG proteins.28, 29, 30, 31 Both IAPs and IAP antagonists are under tight control by various mechanisms to ensure proper regulation of caspase activity.1, 6, 11, 32
Recovery of new drICE alleles as recessive suppressors of GMR-hid. (a) GMR-hid-induced eye ablation phenotype. (b) Heterozygosity of drICE17 does not dominantly modify (suppress) the GMR-hid (Gh) eye ablation phenotype. (c) drICE17homozygously suppresses the GMR-hid (Gh) eye ablation phenotype. Genotype: GMR-hid drICE17(Gh drlCE17)/drICE17. (d) Loss-of-heterozygosity of drICE17 by drICEΔ1 suppresses GMR-hid (Gh). Genotype: GMR-hid drICE17/drICEΔ1. (e–j) drICEL1, drICEL2, drICEC1, drICEC2, drICES1 and drICES2 suppress the GMR-hid (Gh) eye ablation phenotype in trans to drICE17. Genotypes: GMR-hid drICE17/FRT82B drICEX. x denotes the drICE allele. See also Supplementary Figure S1
Given the pivotal roles of apoptosis in development and tissue homeostasis, it is not surprising that deregulation of caspases has been implicated in various pathological conditions, including neurodegeneration, autoimmune diseases and cancers.33 Mutations in Caspase-10 and -8 are found in autoimmune lymphoproliferative syndrome (ALPS) and ALPS-related disorders.34 Mutations and polymorphisms of Caspase-8, -9, -3 and -7 have been implicated in various cancers.35, 36 Although some of these mutations disrupt the apoptotic activity of the affected caspase in cell culture studies,37, 38, 39, 40, 41 detailed understanding of how these mutations affect the function and expression of the caspases is scarce. It is thus of great biological and clinical significance to isolate more caspase mutants, especially the ones carrying point mutations, and to analyze their behaviors in vivo.
The Drosophila model system provides a great venue to answer such inquiries, given the conservation of caspase genes and the well-established genetic techniques to isolate and characterize mutations in genes of interest. DrICE and Dcp-1 are the effector caspase orthologs of mammalian Caspase-3 and Caspase-7 (ref. 42,43; reviewed in ref. 2, 6). Although Dcp-1 has a crucial role in nurse cell death during mid-oogenesis, DrICE is required for apoptosis in most developmental and irradiation-induced cell death.25, 44, 45 Four drICE mutants have been reported so far, only one of which, drICE17, is caused by a missense mutation.25, 44, 46 drICEΔ1, drICEΔ1E4 and drICEΔ2C8 were generated through imprecise P-element excisions, through which the entire drICE open-reading frame was removed to produce null alleles.44, 46 drICE17 carries an N to Y mutation at residue 116 and was isolated in an EMS mutagenesis screen as a recessive suppressor of the small eye phenotype induced by GMR-hid. This missense mutation leads to decreased protein stability and behaves as a strong hypomorphic allele.25 These alleles have improved our understanding of the essential roles of drICE in developmental, stress-induced and RHG-induced cell death;25, 44, 46 however, they have not provided information on how various amino-acid substitutions affect the behavior of the caspase protein.
In this study, we describe the isolation and characterization of six new EMS-induced recessive loss-of-function alleles of drICE, all of which affect highly conserved amino-acid residues. Four of these alleles produce unstable DrICE proteins. However, the remaining two alleles behave genetically differently. Although they were isolated as loss-of-function alleles homozygously, they surprisingly display gain-of-function characteristics in the presence of a wild-type copy of drICE, that is, in a heterozygous condition. Interestingly, they only behave as gain-of-function mutants in the presence of an apoptotic signal. Molecularly, the mutations in these alleles change conserved residues in close proximity to the catalytic Cys residue. We discuss models about how these mutations cause the gain-of-function character of these alleles.
Results
Isolation of de novo drICE mutant alleles
Ectopic expression of hid under the control of the eye-specific GMR promoter causes an eye ablation phenotype owing to excessive cell death (Figure 1a).14 This eye ablation phenotype has been used in EMS mutagenesis screens to isolate mutants of genes involved in apoptosis. To isolate additional drICE mutant alleles to broaden our understanding of DrICE function in cell death, we performed an allele screen. The existing drICE allele, drICE17, does not suppress the GMR-hid-induced eye ablation phenotype in a dominant or heterozygous manner (Figure 1b). Only homozygously (Figure 1c), or in trans to a drICE deficiency (drICEΔ1) (Figure 1d), can suppression of GMR-hid be recorded. Therefore, to isolate new alleles of drICE, we took advantage of the fact that loss-of-heterozygosity of drICE17 causes strong suppression of GMR-hid. In an EMS mutagenesis screen, we screened 40 000 F1 progeny and recovered six suppressors, namely L1, L2, C1, C2, S1 and S2, which suppressed GMR-hid only in trans to drICE17 (Figure 1e–j) and are therefore good candidates for new drICE alleles. In addition, the six mutants are homozygously viable with wing clearance defects (data not shown) as observed in drICE17 and drICEΔ1 alleles.25, 44
Molecular characterization of the novel drICE alleles
Through DNA sequencing, we identified in each of these six alleles point mutations affecting amino acids highly conserved among effector caspases in various species (Figure 2a). drICEL1 and drICEL2 contain missense mutations of amino acids located in the large subunit, changing Y86 to N and G94 to D, respectively. drICES1 and drICES2 bear nonsense mutations in the small subunit at positions 258 and 266, respectively, and produce carboxyl-end truncated drICE proteins. drICEC1 and drICEC2 carry missense mutations of amino acids located near the catalytic cysteine, C211, changing G213 to D and G219 to E, respectively. We refer to the alleles affecting the large subunit as class L alleles (L1 and L2), except the ones located close to the catalytic cysteine residue, which we refer to as class C alleles (C1 and C2). Mutants affecting the small subunit are class S alleles (S1 and S2) (Figure 2b).
The new drICE alleles affect highly conserved amino acids. (a) Alignment of various effector caspases in insects, mammals and humans. drICEL1 and drICEL2 carry Y86N and G94D missense mutations, respectively. drICEC1 and drICEC2 mutants bear G213D and G219E missense mutations, respectively. drICES1 and drICES2 carry nonsense mutations at W258 and W266, respectively. As previously published, drICE17changes N116 to Y.25 (b) Schematic outline of DrICE protein and the relative locations of the mutations in DrICE. Class L alleles carry missense mutations affecting conserved amino acids in the large subunit, which includes drICEL1, drICEL2 and drICE17. Class S alleles are drICES1 and drICES2, both of which carry nonsense mutations in the small subunit. The class C group is composed of drICEC1 and drICEC2, both of which carry missense mutation affecting amino acids in the vicinity of the catalytic cysteine residue
DrICE protein levels are markedly decreased in class L and class S mutants, but not in class C alleles
To assess how these novel drICE mutations affect DrICE function, we examined the stability of the mutant DrICE proteins. Mosaic eye imaginal discs of the drICE alleles from third instar larvae were labeled with an anti-DrICE polyclonal antibody.47 Levels of DrICE protein decreased markedly in mutant clones of class L and class S alleles (Figure 3a’–e’), similar to drICE17, which was previously shown to encode an unstable DrICE protein (ref. 25). These results suggest that class L and class S alleles produce little or unstable DrICE protein.
DrICE protein distribution in drICE mutant clones. Confocal scannings of immunofluorescence images of third instar larval mosaic eye imaginal discs are shown. (a–g) Mutant drICE clones for the indicated alleles are marked by the absence of GFP (green). (a’–g’) anti-DrICE labeling is shown in red. (a–e) Class L and S alleles produce little or unstable DrICE protein. Please note that the anti-DrICE labeling in (a’–e’) matches the outline of the GFP labeling in (a–e), which marks wild-type or heterozygous tissue. Genotypes: ey-FLP; FRT82B drICEX /FRT82B ubi-GFP with x indicating the mutant drICE allele. (f, f’, g, g’) DrICE protein levels are not changed or only slightly decreased in class C mutant clones. Genotypes: (f, f’) ey-FLP; FRT82B drICEC1/FRT82B ubi-GFP; (g, g’) ey-FLP; FRT82B drICEC2/FRT82B ubi-GFP
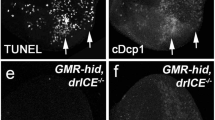
In contrast, class C alleles behave differently. Although mutant clones of drICEC1 contain slightly decreased protein levels, drICEC2 produces normal DrICE protein levels comparable to wild-type tissues (Figure 3f’ and g’). To verify that the class C alleles indeed encode recessive loss-of-function alleles, we examined if they affect normal developmental cell death during eye development. At 28 h after puparium formation (APF), a wave of apoptosis removes all excess interommatidial cells.48, 49, 50 This developmental cell death is lost in mutant clones of both class C alleles (Figure 4a’ and b’), suggesting that class C alleles are recessive loss-of-function mutants. Therefore, because these two alleles produce normal DrICE protein levels, they affect DrICE activity independently of protein stability. Given that they have mutations in highly conserved residues close to the catalytic cysteine residue (Figure 2), it is likely that these mutants have reduced catalytic activity.
Developmental cell death in the pupal retina is absent in mutant clones of the class C alleles. drICEC1 (a) and drICEC2 (b) mosaic pupal eye discs 28 h after puparium formation (APF). drICE mutant clones are marked by absence of GFP (see dashed lines as examples). (a’,a’’,b’,b’’) TUNEL labeling indicates the absence of developmental cell death in C class drICE mutant clones
Class C alleles affect substrate binding
From a structural perspective, it is quite clear why these mutations (G213D and G219E) lead to inactivation of DrICE when they are homozygous. In caspases generally, the substrate-binding groove loops 2, 3 and 4 from one half of the tetramer interact with loop 2′ from the opposite half of the tetramer to form an ordered substrate-binding groove (Figure 5a). Loops 2 and 2′ form a lock that holds loops 3 and 4 into the proper conformation. If the loop 2/loop 2′ interaction is lost, loops 3 and 4 become disordered and the enzyme is unable to bind substrate (Figure 5c). Based on examination of the DrICE structure (3SIP),51 residues G213 and G219 are both contained within loop 2 of DrICE. Neither of these positions can accommodate any residue larger than a glycine (Figure 5b) and the G213D and G219E mutations are predicted to inhibit critical interactions between loops 2 and 2’. G213 forms an exceptionally tight contact with the backbone of F256 on loop 3, the loop that forms the base of the substrate-binding groove (Figure 5b). Substitution of G213 with aspartate disrupts the contact with F256 and prevents the tight association of loops 2 and 2′, which supports substrate binding. G219, also on loop 2, interacts with the side chain of V241, which is contained on loop 2′ (Figure 5b). Substitution of G219 with any rotomers of glutamate results in steric clash with adjacent residues. This steric clash would be sufficient to prevent loop ordering and substrate binding (Figure 5c), explaining the loss of enzymatic activity of these two mutants in a homozygous condition.
Loop position explains loss of activity in homozygous class C mutants. (a) Shown is a schematic drawing of a wild-type DrICE tetramer. The two large subunits of the tetramer are illustrated in light blue, the small subunits in dark blue behind the large subunits. The substrate-binding groove loops 2, 3 and 4 from one half of the tetramer and 2′ from the other half are indicated. The position of substrate (pink rods) is modeled based on superposition of DrICE with the highest available resolution caspase-3 structure (2CNN).67, 68 The substrate binds in the same region as the inhibitory region of the DIAP1 present in the DrICE (3SIP) structure.51 (b) Based on examination of the DrICE structure (3SIP), residues G213 and G219 are both contained within loop 2 of DrICE. G213 forms an exceptionally tight contact with the backbone of F256 on loop 3, the loop that forms the base of the substrate-binding groove. G219 interacts with the side chain of V241, which is contained on loop 2’. At both positions, 213 and 219, any residue larger than glycine would sterically clash and lead to destabilization or distortion of the active form of the enzyme. The G213D and G219E mutations are predicted to prevent the tight association of loops 2 and 2’, which supports substrate binding. (c) A homozygous class C DrICE tetramer. Steric clash of G213D and G219E with residues on the L2’ loop prevents the necessary interactions between loops 2 and 2’ and thus prevents the proper conformation of all four of the substrate-binding groove loops (compare with a). (d) Wild-type/class C DrICE heterotetramer contains one functional and one non-functional DrICE dimer (compare with a)
Class C drICE alleles dominantly enhance GMR-reaper
The new drICE alleles were recovered as recessive suppressors of GMR-hid (Figure 1). They are also recessive suppressors of GMR-reaper (Supplementary Figure S1). Surprisingly, however, in the course of this analysis, we noted that the two class C alleles—when heterozygous in trans to a wild-type (drICE+) allele—dominantly enhanced GMR-reaper (Figure 6b), whereas class L and class S alleles weakly suppress it (Figure 6a; Supplementary Figure S2). Class C alleles also appear to act as dominant enhancers of GMR-hid (Supplementary Figure S3).
Class C alleles enhance the GMR-reaper-induced eye ablation phenotype. (a, b) The class L allele drICEL1 suppresses the GMR-reaper-induced eye ablation phenotype (a) compared with GMR-reaper alone. (b) More examples for the effect of class L and class S alleles on GMR-reaper are shown in Supplementary Figure S2. (c, d) The class C alleles drICEC1 and drICEC2 dominantly enhance the GMR-reaper-induced eye ablation phenotype. (e) Fluorometric caspases assays with head extracts from flies of the indicated genotype. Substrate (DEVD-AMC) cleavage activity of head extracts is presented as relative fluorescence units (RFU) with change over time. (f–i) TUNEL labelings of GMR-reaper eye discs in wild-type or heterozygous drICE mutant background. Although the class L allele drICEL1heterozygously decreases the number of TUNEL-positive nuclei compared with GMR-reaper alone (f), class C alleles increase it (g, h). (j) Statistical summary of (f–i). The two-tailored student’s t-test was used. P-values are indicated. Genotypes: (a, e) CyO,2xGMR-reaper/+; FRT82B drICEL1 / +. (b, f) CyO,2xGMR-reaper/+; FRT82B/ +. (c, g) CyO,2xGMR-reaper/+; FRT82B drICEC1 / +. (d, h) CyO,2xGMR-reaper/+; FRT82B drICEC2/+. See also Supplementary Figures S2 and S3
One possibility by which class C alleles dominantly enhance GMR-reaper is through increased caspase activity. To examine this possibility, we performed fluorometric caspase assays with head extracts from GMR-reaper animals heterozygous for various drICE alleles. Consistently, although the loss-of-function alleles drICEL2 and drICES1 have lost significant caspase activity, the class C allele drICEC1 displayed increased caspase activity compared with GMR-reaper alone (Figure 6e). Furthermore, more TUNEL-positive signals are observed in GMR-reaper eye imaginal discs heterozygous for class C alleles compared with GMR-reaper in wild-type or heterozygous class L background (Figure 6f–j). Therefore, the dominant enhancement of the eye ablation phenotype of GMR-reaper by class C alleles is indeed due to increased cell death.
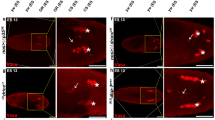
To further characterize the new drICE alleles, we examined DIAP1 degradation triggered by reaper.16, 17, 18, 19, 20 In GMR-reaper eye imaginal discs, DIAP1 degradation is very prominent in cells immediately posterior to the column of R8 photoreceptor neurons, which are very resistant to apoptosis52 and where DIAP1 is not degraded (Figure 7a and b; R8 photoreceptor columns are marked by arrows; the zones of DIAP1 degradation by asterisks (*)). Interestingly, reduced DrICE activity in heterozygous loss-of-function alleles partially protects DIAP1 from reaper-induced degradation (Figure 7c and d), which is consistent with the suppression of GMR-reaper by these alleles (Figure 6a). In contrast, the gain-of-function alleles drICEC1 and drICEC2 fail to protect DIAP1 from reaper-induced degradation (asterisks in Figure 6e and f). These findings suggest that the complex interaction between reaper, DIAP1 and DrICE is affected by the class C alleles.
Reaper-induced DIAP1 degradation is differently influenced by drICE alleles. The white arrow indicates the column of R8 photoreceptor neurons. The zone of DIAP1 degradation is marked by the asterisk. Posterior is to the right. (a) Anti-DIAP1 labeling of a wild-type eye imaginal disc. (b) Anti-DIAP1 labeling of a GMR-reaper eye imaginal disc. Although DIAP1 protein persist in R8 photoreceptor neurons (white arrow), the cells immediately posterior to it strongly degrade DIAP1 (c, d). The loss-of-function alleles drICEL2 and drICES1 partially protect DIAP1 from reaper-induced degradation. The zone of DIAP1 degradation is partially restored (e, f). The gain-of-function alleles drICEC1 and drICEC2 fail to protect DIAP1 from reaper-induced degradation
We also examined if the class C alleles behaved as gain-of-function alleles in the absence of an apoptotic signal such as GMR-reaper, and thus may cause inappropriate cell loss using the developing retina as a model. At 42 h APF, the retina forms a highly regular lattice with a constant number of cells48, 49, 50 (Supplementary Figure S4). Inappropriate cell loss results in disruption of lattice symmetry and is easy to score. However, for both class C alleles, we did not detect any irregularity in the appearance of the lattice that may indicate inappropriate cell loss (Supplementary Figure S4). This analysis suggests that the class C mutations do not cause premature and inappropriate activation of DrICE in the absence of an apoptotic signal.
Taken together, the class C alleles exert a very complex genetic behavior. Homozygously mutant drICEC1 and drICEC2 are recessive loss-of-function alleles and produce mutant DrICE proteins with impaired substrate binding. However, in the presence of functional DrICE protein encoded from the drICE+ allele, they trigger more cell death than normal and enhance the GMR-reaper eye ablation phenotype. Thus, in a heterozygous condition they behave as gain-of-function alleles.
Discussion
In this study, we report the isolation and characterization of six new mutants of the Drosophila effector caspase drICE, the ortholog of mammalian caspase-3. According to the types and locations of the mutations in these six new drICE alleles, the expression levels of the mutant DrICE proteins and the genetic interactions with GMR-hid or GMR-reaper, we have grouped these mutants into three classes. Class L and S alleles carry point mutations in the large and small subunits, respectively (Figure 2b). In addition, according to this classification criterion, drICE17(N116Y) also belongs to the class L group. Both class L and class S alleles affect the stability of their DrICE protein products and hence have reduced caspase activity. Overall, they are recessive loss-of-function drICE alleles.
The class C alleles drICEC1 and drICEC2 carry missense mutations in conserved amino acids (G213D and G219E) in close proximity of the catalytic cysteine (C211) residue. In contrast to class L and class S alleles, these mutations do not markedly impact DrICE protein stability. Most importantly, although these alleles act as loss-of-function alleles homozygously, in the presence of a wild-type drICE+ allele (i.e., heterozygously), they can enhance cell death induced by reaper expression, suggesting that they can function as gain-of-function alleles. However, our analysis suggests that class C mutants only behave as gain-of-function alleles in an apoptotic background such as GMR-reaper, whereas they do not cause spontaneous activation of DrICE in the absence of apoptotic signals. Therefore, they can only act as gain-of-function mutants after proteolytic processing.
The underlying mechanisms of the apoptotic enhancement by heterozygous class C alleles are less clear. In the few cases where certain mutants behave as recessive loss-of-function and dominant gain-of-function alleles, dimerization is the underlying cause of this different genetic behavior. For example, certain mutations of the Toll receptor are recessive loss-of-function alleles, but are gain-of-function owing to physical interaction with the wild-type allele.53, 54 Given that two processed DrICE molecules form an enzymatically active tetramer with two catalytic sites,2, 3 one can speculate that class C mutant subunits exert a dominant effect via interaction with a wild-type DrICE molecule. Statistically, only 50% of the DrICE tetramers in the cell are composed of a wild-type and a class C subunit (referred to as wt/class C heterotetramer; Figure 5d). The remaining 50% are either wild-type (Figure 5a) or class C tetramers (Figure 5c), the latter of which has no enzymatic activity. Thus, the wt/class C heterotetramers actually have more activity as the data imply because they also compensate for the loss of activity of the class C tetramers.
How can we explain the gain-of-function activity of the wt/class C heterotetramers? Because of the loss of loop organization and substrate binding in the class C subunit (Figure 5), it is unlikely that the presence of the wild-type subunit in the wt/class C heterotetramer 'rescues' the defect in the class C subunit. It is more likely that an upstream regulatory mechanism is deregulated by the wt/class C heterotetramer. There are several possibilities. For example, because IAPs require effector caspases for inhibitory activity,55, 56 it may be possible that a wt/class C heterotetramer does not produce as much active DIAP1 compared with a wild-type tetramer. Specifically, the N terminus of DIAP1 is an intramolecular inhibitor of DIAP1, keeping DIAP1 in an auto-inhibited conformation, unable to bind and inhibit DrICE.51, 57, 58, 59 After caspase cleavage at D20 (likely by DrICE), the inhibitory N terminus is removed and now activated DIAP1 can bind to DrICE and inhibit it. Because DIAP1 is a substrate of DrICE before it becomes an inhibitor, the wt/class C heterotetramer would produce less-active DIAP1 and thus would be less-efficiently inhibited.
Another possible model includes cooperativity of the binding of DIAP1 to the two active sites of the DrICE tetramer. Because the inhibitory region of DIAP1 (pink rod in Figure 5d) binds to the substrate-binding domain of DrICE present in the 3SIP structure,51 DIAP1 does not bind to the mutant catalytic site of the class C subunit. If there is cooperativity of the binding of DIAP1 to the two catalytic sites of DrICE, the failure of DIAP1 to bind to the class C catalytic side may affect the binding of DIAP1 to the catalytic site of the wild-type subunit. Thus, the catalytic site of the wild-type subunit would be free from IAP inhibition, which should lead to increased activity only in the heterozygous condition.
Finally, we also take into account that class C alleles are only dominantly active in the presence of an apoptotic signal such as reaper (Figure 6,Supplementary Figure S4). Reaper and DrICE compete for binding to the BIR1 domain of DIAP1.47 If reaper is in excess, it triggers degradation of DIAP1 (Figure 7b) and thus apoptosis.16, 17, 18, 19, 20 Therefore, owing to the failure of DIAP1 to bind to the class C subunit, the wt/class C heterotetramer may shift the equilibrium towards binding of DIAP1 to reaper such that DIAP1 is more efficiently degraded by reaper, resulting in higher enzymatic activity of the wt/class C heterotetramer. Consistent with this notion, reduced DrICE activity in heterozygous loss-of-function alleles partially protects DIAP1 from reaper-induced degradation (Figure 7c and d), whereas the class C alleles of DrICE fail to protect DIAP1 from degradation (Figure 7e and f), suggestive of decreased binding of the wt/class C heterotetramer to DIAP1.
These models to explain the gain-of-function behaviors of the class C alleles are not mutually exclusive and other models may be possible, too.
To our knowledge, this is the first time that viable gain-of-function alleles of caspases are described in any intact organism and provides a significant exception to the expectation that mutations of conserved amino acids always abolish the pro-apoptotic activity of caspases. Because the affected residues of the class C alleles are conserved in other caspases (Figure 2a), it would be of great interest to generate and test such mutations in other caspases to examine if it is a universal phenomenon. In Caenorhabditis elegans, a large collection of ced-3 mutants has been characterized, but no gain-of-function alleles have been reported.60 Interestingly, one ced-3 allele, n2433, substitutes G360 with S. This glycine residue corresponds to G213 in DrICE, which is changed to D in drICEC1 (Figure 2). However, in contrast to drICEC1, n2433 behaves as a dominant negative.60 It is unclear, if these different genetic behaviors are due to the different amino-acid substitutions (S in n2433 vs D in drICEC1) or to intrinsic differences between Ced-3 and DrICE.
In mice, the Melody mutation in Caspase-3 substitutes the catalytic C with a S residue. This mutation also behaves as a dominant negative in a heterozygous condition.61 The only mutation in Caspase-3 that confers constitutively active properties to Caspase-3 is the V266E substitution, which was characterized by in vitro mutagenesis.62, 63 This mutation increases Caspase-3 activity 60-fold. V266 is located in the dimer interface of the small subunit and the V266E mutation promotes dimerization of Caspase-3 without proteolytic processing.63 V266 is not conserved in DrICE or other caspases (Figure 2), but even if it was, it is unlikely that such a mutation can be recovered in vivo as it will cause dominant lethality due to excessive apoptosis.
The allele-specific enhancement of apoptotic activity by caspase mutants is also of clinical significance. For example, it might be of interest to determine whether such dominant mutations of caspases have a role in the pathogenesis and development of neurodegenerative disorders, for which the underlying mechanism is apoptotic neuron loss. Sequence analysis of caspase genes in these patients will help in answering this possibility. Conversely, it would also be of interest to design and screen for drugs that would slightly twist the conformation of caspase tetramers to increase their enzymatic activity and induce cell death in tumors.
Materials and Methods
Isolation and identification of drICE mutant alleles
An isogenized FRT82B stock was used for EMS mutagenesis and also used as a control for genetic analyses. FRT82B males were treated with 25 mM EMS in 5% sucrose solution for 24 h. After recovery for 3 h, they were crossed to GMR-hid drICE17 females, and incubated at 25 °C. In all, 40 000 F1 progeny were screened for suppression of the small eye phenotype of GMR-hid drICE17. Six mutants were identified as de novo drICE alleles by genetic analysis and DNA sequencing as described in the Results section.
Fly stocks and genetics
The following mutant and transgenic fly stocks were used: drICEL1; drICEL2; drICEC1; drICEC2; drICES1; drICES2 (this study); drICE17 (ref. 25); drICEΔ1 (ref. 44); GMR-hid;14 CyO,2xGMR-reaper;12 ey-FLP, FRT82B ubi-GFP;64 GMR-hid ey-FLP, FRT82B ubi-GFP.25 GMR-hid drICE17 is a recombinant chromosome carrying a GMR-hid transgene and drICE17. Genetic mosaics for immunohistochemical analysis were obtained by crossing the FRT82B drICE alleles with ey-FLP; FRT82B drICE-/FRT82B ubi-GFP.
DNA sequencing
Genomic DNA from homozygous mutant flies or trans-heterozygous over drICEΔ1flies was isolated and PCR-amplified using drICE-specific primers. PCR fragments were sequenced by Sanger sequencing. The single amino-acid code was used.
Immunohistochemistry
TUNEL and immunohistochemistry were carried out as described.65 Anti-DrICE antibody (a kind gift of Pascal Meier) was raised in guinea pig47 and used at a dilution of 1 : 200. Anti-Dlg antibody (a kind gift of Georg Halder) was used at a dilution of 1 : 2000. Anti-DIAP1 (SK14) antibody (a kind gift of Pascal Meier) was raised in guinea pig and used at a concentration of 1 : 400. Cy3- and Cy-5 fluorescent-conjugated secondary antibodies were obtained from Jackson ImmunoResearch (West Grove, PA, USA). In general, 10–20 eye imaginal discs were analyzed, unless stated otherwise. Fluorescent Images were captured using a Zeiss Axio Imager Z1 with ApoTome technology or an Olympus Optical FV500 confocal microscope.
Quantification of GMR-reaper-induced excessive cell death
All TUNEL-positive cells in GMR-reaper larval eye discs in various genotypes were counted to indicate how much cell death was induced. For each genotype, 15–17 representative eye discs were counted. Significance was calculated by using unpaired two-tailed Student’s t-test with a 95% confidence interval.
Fluorometric caspase assays
Fluorometric caspase assays were performed as described with modifications.66 Adult heads were dissected on ice and lysed with a pestle in caspase assay buffer (50 mM HEPES, pH 7.5; 100 mM NaCl; 1 mM EDTA; 0.1% CHAPS; 10% sucrose; 5 mMDTT; 0.5% TritonX‐100; 4% glycerol; 1 × protease inhibitor cocktail (Promega, Madison, WI, USA)) and then sonicated once for 7 s. Protein concentrations of the lysates were determined by Bradford Protein Assay. For each genotype, 40 μg of total protein lysates were used, respectively. Protein lysates and 100 μM of the fluorometric substrate DEVD-AMC (MP Biomedicals, Santa Ana, CA, USA) were combined in a 96-well plate on ice. Reaction volume was brought to 100 μl with caspase assay buffer. Spectrophotometer was used to measure fluorescence (excitation 385 nm and emission 460 nm) at 15 min intervals for 2 h at 37 °C.
Abbreviations
- ALPS:
-
autoimmune lymphoproliferative syndrome
- APF:
-
after puparium formation
- Ced-3:
-
cell death defective 3
- CyO:
-
curly of Oster
- Dcp-1:
-
death caspase 1
- DIAP1:
-
death-associated inhibitor of apoptosis 1
- DNA:
-
desoxyribonucleic acid
- drICE:
-
death-related ICE (interleukin converting enzyme)
- dronc:
-
death regulator Nedd2-like caspase
- EMS:
-
ethyl methanesulfonate
- Ey:
-
eyeless
- Flp:
-
flippase
- FRT82B:
-
flippase recombination target at 82B
- GFP:
-
green fluorescent protein
- Gh:
-
GMR-hid
- Hid:
-
head involution defective
- GMR:
-
glass multimer reporter
- HtrA2:
-
high-temperature-required protein A2
- IAP:
-
inhibitor of apoptosis protein
- PCR:
-
polymerase chain reaction
- R8:
-
photoreceptor 8
- RHG:
-
reaper, hid, grim
- Smac/Diablo:
-
second mitochondria-derived activator of caspases/direct IAP binding protein with low pI
- TUNEL:
-
terminal deoxynucleotidyl transferase dUTP nick end labeling
- Ubi:
-
ubiquitous
- wt:
-
wild-type
- XIAP:
-
X-linked inhibitor of apoptosis
References
Fuchs Y, Steller H . Programmed cell death in animal development and disease. Cell 2011; 147: 742–758.
Kumar S . Caspase function in programmed cell death. Cell Death Differ 2007; 14: 32–43.
Riedl SJ, Shi Y . Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol 2004; 5: 897–907.
Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW et al. Human ICE/CED-3 protease nomenclature. Cell 1996; 87: 171.
Bratton SB, Salvesen GS . Regulation of the Apaf-1-caspase-9 apoptosome. J Cell Sci 2010; 123 (Pt 19): 3209–3214.
Xu D, Woodfield SE, Lee TV, Fan Y, Antonio C, Bergmann A . Genetic control of programmed cell death (apoptosis) in Drosophila. Fly 2009; 3: 78–90.
Callus BA, Vaux DL . Caspase inhibitors: viral, cellular and chemical. Cell Death Differ 2007; 14: 73–78.
Vaux DL, Silke J . IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol 2005; 6: 287–297.
Gyrd-Hansen M, Meier P . IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer 2010; 10: 561–574.
Galban S, Duckett CS . XIAP as a ubiquitin ligase in cellular signaling. Cell Death Differ 2010; 17: 54–60.
Orme M, Meier P . Inhibitor of apoptosis proteins in Drosophila: gatekeepers of death. Apoptosis 2009; 14: 950–960.
White K, Tahaoglu E, Steller H . Cell killing by the Drosophila gene reaper. Science 1996; 271: 805–807.
Chen P, Nordstrom W, Gish B, Abrams JM . grim, a novel cell death gene in Drosophila. Genes Dev 1996; 10: 1773–1782.
Grether ME, Abrams JM, Agapite J, White K, Steller H . The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev 1995; 9: 1694–1708.
White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H . Genetic control of programmed cell death in Drosophila. Science 1994; 264: 677–683.
Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H . Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol 2002; 4: 432–438.
Holley CL, Olson MR, Colon-Ramos DA, Kornbluth S . Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat Cell Biol 2002; 4: 439–444.
Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL et al. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol 2002; 4: 416–424.
Hays R, Wickline L, Cagan R . Morgue mediates apoptosis in the Drosophila melanogaster retina by promoting degradation of DIAP1. Nat Cell Biol 2002; 4: 425–431.
Wing JP, Schreader BA, Yokokura T, Wang Y, Andrews PS, Huseinovic N et al. Drosophila Morgue is an F box/ubiquitin conjugase domain protein important for grim-reaper mediated apoptosis. Nat Cell Biol 2002; 4: 451–456.
Hay BA, Wassarman DA, Rubin GM . Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 1995; 83: 1253–1262.
Goyal L, McCall K, Agapite J, Hartwieg E, Steller H . Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J 2000; 19: 589–597.
Lisi S, Mazzon I, White K . Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics 2000; 154: 669–678.
Xu D, Li Y, Arcaro M, Lackey M, Bergmann A . The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development 2005; 132: 2125–2134.
Xu D, Wang Y, Willecke R, Chen Z, Ding T, Bergmann A . The effector caspases drICE and dcp-1 have partially overlapping functions in the apoptotic pathway in Drosophila. Cell Death Differ 2006; 13: 1697–1706.
Srivastava M, Scherr H, Lackey M, Xu D, Chen Z, Lu J et al. ARK, the Apaf-1 related killer in Drosophila, requires diverse domains for its apoptotic activity. Cell Death Differ 2007; 14: 92–102.
Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J et al. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol 2002; 4: 445–450.
Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE et al. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 2000; 102: 43–53.
Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R . A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell 2001; 8: 613–621.
Du C, Fang M, Li Y, Li L, Wang X . Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 2000; 102: 33–42.
Hegde R, Srinivasula SM, Zhang Z, Wassell R, Mukattash R, Cilenti L et al. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J Biol Chem 2002; 277: 432–438.
Bergmann A . The role of ubiquitylation for the control of cell death in Drosophila. Cell Death Differ 2010; 17: 61–67.
Agostini M, Tucci P, Melino G . Cell death pathology: perspective for human diseases. Biochem Biophys Res Commun 2011; 414: 451–455.
Madkaikar M, Mhatre S, Gupta M, Ghosh K . Advances in autoimmune lymphoproliferative syndromes. Eur J Haematol 2011; 87: 1–9.
Ghavami S, Hashemi M, Ande SR, Yeganeh B, Xiao W, Eshraghi M et al. Apoptosis and cancer: mutations within caspase genes. J Med Genet 2009; 46: 497–510.
Olsson M, Zhivotovsky B . Caspases and cancer. Cell Death Differ 2011; 18: 1441–1449.
Kim MS, Kim HS, Jeong EG, Soung YH, Yoo NJ, Lee SH . Somatic mutations of caspase-2 gene in gastric and colorectal cancers. Pathol Res Pract 2011; 207: 640–644.
Park WS, Lee JH, Shin MS, Park JY, Kim HS, Kim YS et al. Inactivating mutations of the caspase-10 gene in gastric cancer. Oncogene 2002; 21: 2919–2925.
Kim HS, Lee JW, Soung YH, Park WS, Kim SY, Lee JH et al. Inactivating mutations of caspase-8 gene in colorectal carcinomas. Gastroenterology 2003; 125: 708–715.
Soung YH, Lee JW, Kim HS, Park WS, Kim SY, Lee JH et al. Inactivating mutations of CASPASE-7 gene in human cancers. Oncogene 2003; 22: 8048–8052.
Soung YH, Lee JW, Kim SY, Sung YJ, Park WS, Nam SW et al. Caspase-8 gene is frequently inactivated by the frameshift somatic mutation 1225_1226delTG in hepatocellular carcinomas. Oncogene 2005; 24: 141–147.
Fraser AG, Evan GI . Identification of a Drosophila melanogaster ICE/CED-3-related protease, drICE. EMBO J 1997; 16: 2805–2813.
Song Z, McCall K, Steller H . DCP-1, a Drosophila cell death protease essential for development. Science 1997; 275: 536–540.
Muro I, Berry DL, Huh JR, Chen CH, Huang H, Yoo SJ et al. The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development 2006; 133: 3305–3315.
Laundrie B, Peterson JS, Baum JS, Chang JC, Fileppo D, Thompson SR et al. Germline cell death is inhibited by P-element insertions disrupting the dcp-1/pita nested gene pair in Drosophila. Genetics 2003; 165: 1881–1888.
Kondo S, Senoo-Matsuda N, Hiromi Y, Miura M . DRONC coordinates cell death and compensatory proliferation. Mol Cell Biol 2006; 26: 7258–7268.
Zachariou A, Tenev T, Goyal L, Agapite J, Steller H, Meier P . IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. EMBO J 2003; 22: 6642–6652.
Baker NE . Cell proliferation, survival, and death in the Drosophila eye. Semin Cell Dev Biol 2001; 12: 499–507.
Brachmann CB, Cagan RL . Patterning the fly eye: the role of apoptosis. Trends Genet 2003; 19: 91–96.
Roignant JY, Treisman JE . Pattern formation in the Drosophila eye disc. Int J Dev Biol 2009; 53: 795–804.
Li X, Wang J, Shi Y . Structural mechanisms of DIAP1 auto-inhibition and DIAP1-mediated inhibition of drICE. Na Commun 2011; 2: 408.
Fan Y, Bergmann A . Multiple mechanisms modulate distinct cellular susceptibilities toward apoptosis in the developing Drosophila eye. Dev Cell 2014; 30: 48–60.
Anderson KV, Jurgens G, Nusslein-Volhard C . Establishment of dorsal-ventral polarity in the Drosophila embryo: genetic studies on the role of the Toll gene product. Cell 1985; 42: 779–789.
Schneider DS, Hudson KL, Lin TY, Anderson KV . Dominant and recessive mutations define functional domains of Toll, a transmembrane protein required for dorsal-ventral polarity in the Drosophila embryo. Genes Dev 1991; 5: 797–807.
Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J et al. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 2001; 410: 112–116.
Tenev T, Zachariou A, Wilson R, Ditzel M, Meier P . IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat Cell Biol 2005; 7: 70–77.
Yan N, Wu JW, Chai J, Li W, Shi Y . Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nat Struct Mol Biol 2004; 11: 420–428.
Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol 2003; 5: 467–473.
Yokokura T, Dresnek D, Huseinovic N, Lisi S, Abdelwahid E, Bangs P et al. Dissection of DIAP1 functional domains via a mutant replacement strategy. J Biol Chem 2004; 279: 52603–52612.
Shaham S, Reddien PW, Davies B, Horvitz HR . Mutational analysis of the Caenorhabditis elegans cell-death gene ced-3. Genetics 1999; 153: 1655–1671.
Parker A, Hardisty-Hughes RE, Wisby L, Joyce S, Brown SD . Melody, an ENU mutation in Caspase 3, alters the catalytic cysteine residue and causes sensorineural hearing loss in mice. Mamm Genome 2010; 21: 565–576.
Pop C, Feeney B, Tripathy A, Clark AC . Mutations in the procaspase-3 dimer interface affect the activity of the zymogen. Biochemistry 2003; 42: 12311–12320.
Walters J, Pop C, Scott FL, Drag M, Swartz P, Mattos C et al. A constitutively active and uninhibitable caspase-3 zymogen efficiently induces apoptosis. Biochem J 2009; 424: 335–345.
Newsome TP, Asling B, Dickson BJ . Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 2000; 127: 851–860.
McCall K, Peterson JS . Detection of apoptosis in Drosophila. Methods Mol Biol 2004; 282: 191–205.
Denton D, Mills K, Kumar S . Methods and protocols for studying cell death in Drosophila. Methods Enzymol 2008; 446: 17–37.
Ganesan R, Jelakovic S, Campbell AJ, Li ZZ, Asgian JL, Powers JC et al. Exploring the S4 and S1 prime subsite specificities in caspase-3 with aza-peptide epoxide inhibitors. Biochemistry 2006; 45: 9059–9067.
Ganesan R, Mittl PR, Jelakovic S, Grutter MG . Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis. J Mol Biol 2006; 359: 1378–1388.
Acknowledgements
We are thankful to Georg Halder (Katholieke Universiteit Leuven, Belgium), Bruce Hay (Caltech, USA), and Pascal Meier (Institute of Cancer Research, UK) for fly stocks and antibodies. This work was supported by the National Institute of General Medical Science (NIGMS) to AB.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by DL Vaux
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Wu, Y., Lindblad, J., Garnett, J. et al. Genetic characterization of two gain-of-function alleles of the effector caspase DrICE in Drosophila. Cell Death Differ 23, 723–732 (2016). https://doi.org/10.1038/cdd.2015.144
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2015.144