Abstract
Activities displaying caspase cleavage specificity have been well documented in various plant programmed cell death (PCD) models. However, plant genome analyses have not revealed clear orthologues of caspase genes, indicating that enzyme(s) structurally unrelated yet possessing caspase specificity have functions in plant PCD. Here, we review recent data showing that some caspase-like activities are attributable to the plant subtilisin-like proteases, saspases and phytaspases. These proteases hydrolyze a range of tetrapeptide caspase substrates following the aspartate residue. Data obtained with saspases implicate them in the proteolytic degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) during biotic and abiotic PCD, whereas phytaspase overproducing and silenced transgenics provide evidence that phytaspase regulates PCD during both abiotic (oxidative and osmotic stresses) and biotic (virus infection) insults. Like caspases, phytaspases and saspases are synthesized as proenzymes, which are autocatalytically processed to generate a mature enzyme. However, unlike caspases, phytaspases and saspases appear to be constitutively processed and secreted from healthy plant cells into the intercellular space. Apoplastic localization presumably prevents enzyme-mediated protein fragmentation in the absence of PCD. In response to death-inducing stimuli, phytaspase has been shown to re-localize to the cell interior. Thus, plant PCD-related proteases display both common (D-specific protein fragmentation during PCD) and distinct (enzyme structure and activity regulation) features with animal PCD-related proteases.
Similar content being viewed by others
Main
Plants, like animals, employ programmed cell death (PCD) for a variety of purposes including development (e.g., xylem formation, seed germination and senescence), stress responses (e.g., salt, cold, heat) and defence (e.g., hypersensitive response (HR)). Although it is becoming increasingly evident that multiple forms of PCD exist in both plants and animals,1, 2 certain forms of PCD in both kingdoms share a number of hallmarks, such as DNA fragmentation, cytochrome c release from mitochondria, cell shrinkage, and so on.3 However, despite extensive similarities, the equivalence of the molecular mechanisms regulating plant PCD is less clearly understood. Among the more surprising findings is an absence of caspase orthologues in plants. Because caspases, a family of cysteine-dependent aspartate-specific proteases, are known to have important functions in the initiation and execution of PCD in animals through cleavage of a number of protein targets, their apparent absence in plants poses a dilemma. Either the principle of specific protein fragmentation at aspartate residues does not operate during PCD in plants or the function of caspases is assumed by other protease(s) that are structurally unrelated to caspases.
Evidence accumulated during the past decade indicates that, in at least some cases, the second scenario is correct and that caspase-like (in a functional rather than a structural sense) proteases in plants have substrate cleavage specificities similar to those of caspases (i.e., cleavage of a peptide bond following an aspartate residue, D, preceded by a specific amino-acid recognition motif). Indeed, synthetic tetrapeptide aldehydes (XXXD-CHO) developed as inhibitors based on the recognition motifs of specific members of the caspase family have repeatedly been reported to prevent PCD in plants.4, 5 Furthermore, several protein inhibitors of caspases (such as p35 of baculovirus and IAP) when expressed in plants have been shown to inhibit the execution of plant PCD.6 Consistent with these findings, protease activities capable of hydrolyzing caspase substrates (the above-mentioned recognition peptides linked to a fluorogenic group at their D residue) have been detected in various plant PCD models and tentatively named VADase, YVADase, VEIDase, TATDase, and so on – depending on the peptide substrate used for their detection (reviewed by Bonneau et al.5) Interestingly, at least some of these corresponding proteolytic activities have been shown to become markedly enhanced upon induction of PCD. Therefore, a hunt for these plant ‘caspase-like’ proteases is warranted and ongoing.
A surprising outcome of this pursuit so far is the revelation that a majority of the diverse caspase-like substrate cleavage specificities (VEIDase, YVADase, VADase, IETDase, LEHDase, WEHDase, and so on – see Table 1 for relative cleavage efficiencies) observed in plants can be assigned to subtilisin-like protease(s) (subtilases). Subtilases are serine-dependent enzymes (in contrast to cysteine-dependent caspases) and are structurally very different from caspases. It should be noted that in plants and distinct from subtilases, YVADase activity is also displayed by an asparagine (N)-specific vacuolar processing enzyme (VPE), a cysteine-dependent plant protease.7, 8 Likewise, DEVDase activity is exhibited by Arabidopsis PBA1, a proteasome subunit.9 In this review, we will focus on two types of plant aspartate (D)-specific subtilases, each of which displays hydrolytic activity towards a broad range of caspase substrates.
Plant Subtilisin-Like Proteases
Subtilisin-like proteases (subtilases) are serine proteases characterized by a catalytic triad of amino acids, aspartate, histidine and serine.10 The arrangement of theses catalytic residues is shared with the prototypical subtilisin from Bacillus licheniformis. According to the MEROPS classification, subtilases constitute the S8 family within the SB clan of serine proteases.11 Plant subtilases correspond to S8A subtilisin subfamily and form an extensive group of enzymes, whereas S8B (kexin-type) proteins appear to be absent from plants.12 In the Arabidopsis genome, 56 subtilase genes have been annotated and appear to encode functional proteases.12, 13 A typical plant subtilase comprises a predicted secretion signal peptide, a pro-domain and a peptidase domain with a protease-associated domain within (Figure 1).12 The mature protease is formed after removal of the signal peptide and the pro-domain.14 The first structure of a plant subtilase, SlSBT3 from tomato, has been determined recently.15
Schematic representation of subtilisin like proteases exemplified by pre–pro-phytaspase.49 Amino-acid numbering is given for tobacco phytaspase (MEROPS ID S08.150). D149, H220 and S537 are active site residues; N322 is the oxyanion hole residue. His331 is predicted to fix the P1 Asp side chain of a substrate (see below). Note that autocatalytic prodomain cleavage occurs after D117 (arrow), in accord with caspase-like specificity of phytaspase. Saspases42 and rice phytaspases49 are organized similarly
A number of subtilases have been purified from plant tissues and analysed. Some of these enzymes are highly abundant and exhibit broad substrate specificity. Thus, an involvement in general protein turnover has been predicted for these proteins.13 An example of a subtilase that functions in protein degrading is cucumisin, which constitutes up to 10% of the soluble protein in melon fruit.16
In addition to merely degradative functions, plant subtilases may also fulfil highly specific functions in plant development and signalling cascades (reviewed by Rautengarten et al.13 and Schaller17). The Arabidopsis SDD1 subtilase has been shown to act extracellularly in the apoplast of stomatal precursor cells where it may be involved in the generation of signals responsible for stomata density regulation.18 Likewise, the subtilase ALE1 may be involved in the generation of peptide signals that are required for cuticle formation and epidermal differentiation during Arabidopsis embryo development.19 The Arabidopsis SBT1.7 subtilase was shown to mediate mucilage release from seed coats,20 and the C1 subtilisin-like protein from soybean, which displays preference for cleavage after Glu, appears to be involved in proteolysis of the seed storage protein, β-conglycinin.21, 22 The Arabidopsis S1P subtilase has been implicated in fragmentation of the membrane-bound transcription factor AtbZIP1723 and precursors of pectin methylesterase24 and rapid alkalinization growth factor.25 Processing of a phytosulphokine growth factor precursor by the Arabidopsis SBT1.1 subtilase has been reported, although the cleavage site does not match the terminus of the mature growth factor.26 The tomato subtilase P69 has been identified as a pathogenesis-related protein and was shown to be one of several subtilases that are specifically induced following pathogen infection.27 P69 was suggested to process a leucin-rich repeat cell wall protein in virus-infected tomato plants. However, the direct consequences of this processing event for pathogenesis are still unknown.28
Despite their prevalence and potential relevance in the regulation of plant development, our current understanding of subtilase function is still very limited. This paper reviews recent findings implicating involvement of plant subtilisin-like proteins with Asp cleavage specificity in the regulation of PCD. Two independent approaches have produced complementary results.
Saspases from Oat
Victorin, an unusual, cyclized pentapeptide,29, 30 is a host-selective toxin produced by the fungus Cochliobolus victoriae. Victorin production is causal to the ability of the fungus to incite the disease Victoria blight.31 As the reference to ‘toxin’ implies, victorin causes cell death in sensitive plants, but does so in a gene-specific manner. Toxin sensitivity (and Victoria blight susceptibility) is conditioned by a single dominant gene, and in oats, the host where Victoria blight was originally identified, this gene is called Vb. Significantly, in oats it was discovered that the Vb gene is inextricably linked to the Pc2 gene for resistance to the crown rust pathogen, Puccinia coronata.32, 33 This and the recent discovery of victorin sensitivity in Arabidopsis thaliana34 and the characterization of the gene that confers victorin sensitivity as a member of a class of genes commonly associated with plant disease resistance (i.e., encodes an NB-LRR protein)35, 36 indicates that the cell death response evoked by victorin is related/identical to the PCD response associated with defence commonly referred to as the HR.
Victorin-induced cell death exhibits many of the characteristics commonly associated with PCD, including cell shrinkage and collapse,37, 38 chromatin condensation,37 DNA laddering,39, 40 mitochondrial depolarization and permeability transition38, 41 and targeted, substrate-specific proteolysis.39 A principle in vivo target of proteolysis is the ∼55 kDa large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) leading to the formation of an ∼53 kDa product, which is stable in the dark, but completely degraded in the light.39 Characterization of the 53 kDa product revealed that cleavage in Rubisco occurs at the N terminus following lysine 14.39 Given that caspases cleave their substrates following aspartate residues, it is clear that the protease(s) directly modifying Rubisco is not a caspase. However, in addition to the inhibition of proteolysis by the general protease inhibitors E-64 (a cysteine protease inhibitor) and leupeptin (a cysteine and serine protease inhibitor), proteolysis is also inhibited by a variety of caspase-specific inhibitors including Ac-VAD-chloromethylketone (CMK), which is generally considered a pan-caspase inhibitor and Ac-DEVD-CMK, which is more specific for caspase-3.42
The inhibition of Rubisco proteolysis by caspase-specific inhibitors suggested the involvement of caspase-like activity. To investigate this possibility, total soluble protein was prepared from victorin-treated, sensitive oat tissue.42 Analysis by hydrophobic interaction chromatography separated two distinct caspase-like activities. One activity was inhibited by the caspase-3 inhibitor, Ac-DEVD-CMK, and hydrolyzed the corresponding fluorogenic substrate, Z-DEVD-AFC, whereas the second activity was inhibited by the pan-caspase inhibitor, Ac-VAD-CMK, and hydrolyzed the corresponding fluorogenic substrate, Z-VAD-AFC. Significantly, the DEVDase activity was not inhibited by the pan-caspase inhibitor (Ac-VAD-CMK) nor did it hydrolyze the corresponding fluorogenic, pan-caspase substrate (Z-VAD-AFC). Conversely, the VADase activity was not inhibited by the caspase-3 inhibitor (Ac-DEVD-CMK) nor did it cleave the corresponding caspase-3 substrate (Z-DEVD-AFC). In addition, neither activity was inhibited by the general protease inhibitors, E-64 or leupeptin. Cumulatively, these data indicate the participation of at least three distinct protease activities in the processing of Rubisco, including two with caspase-like activity. Because the inhibition of any one of these activities prevents Rubisco proteolysis, the data also indicate that these activities direct Rubisco proteolysis through a protease cascade. This is notably similar to the proteolytic processes directed by caspases.
Because of its apparently higher relative abundance, the caspase-like activity capable of cleaving Z-VAD-AFC (pan-caspase substrate) was chosen for further characterization. This activity ultimately separated into two protein fractions termed SAS-1 and -2, which each migrated with an apparent mass of 84 kDa in SDS-PAGE analysis. Matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) mass spectrometry indicated a mass of 74.5 kDa±1900 Da for both proteins. N-terminal sequence analysis was identical for SAS-1 and -2, with the exception that for SAS-1 an N-terminal threonine was not detected. MALDI-TOF spectral analysis of a partial tryptic digest of each protein showed that they both shared all major peaks in the mass profiles, with the exception of two peaks whose size variation indicated either a single amino-acid difference or a difference in post-translational modification. Both proteins displayed nearly identical activities when incubated in a variety of salts, and indistinguishable pH maxima. In addition, both have essentially identical substrate recognition profiles when tested against a suite of caspase fluorogenic substrates. Consequently, SAS-1 and -2 differ either owing to differences in post-translational modifications or, more likely, are the products of very closely-related members of the same gene family.
SAS-2 was further characterized by sequence analysis of three internal peptides released by tryptic digest. N-terminal and internal sequences all showed extensive homology to a variety of plant subtilisin-like serine proteases with the closest match (77.4% overall identity) to a rice subtilisin-like serine protease (GenBank accession number BAB89803).42 These proteins are expressed as pre–pro-proteins whose prodomains are autocatalytically processed (Figure 1). As might be expected, given the substrate specificity of SAS-2, the closely related rice gene encodes an aspartate residue at the putative prodomain cleavage site. This characterization coupled with the finding that aldehyde and fluoromethyketone (FMK)-based inhibitors inhibit SAS-1 and -2 reversibly and only CMK-based inhibitors irreversibly inhibit the enzymes show that SAS-1 and -2 are serine proteases (FMK- and CMK-based inhibitors irreversibly inhibit cysteine proteases, whereas only CMK inhibitors are irreversible for serine proteases).43 Thus, given that they are serine proteases exhibiting ‘aspase’ activity, and following an equivalent convention of calling cysteine proteases exhibiting ‘aspase’ activity ‘caspases’, these enzymes were called ‘saspases’ (not to be confused with the subsequently named ‘SASPase’ for skin aspartic protease44).
Like caspases, the saspases show substrate specificity beyond simply requiring aspartate at the P1 residue.42 Of the 10 caspase-based substrates tested, they do not cleave the preferred substrates for caspase-2, -3, -5 and -6 (based on the peptide recognition sequences: ‘VDVAD’, ‘DEVD’, ‘WEHD’ and ‘VEID’, respectively) and are only marginally active against the preferred substrates for caspase-1 and -4 (based on the peptide recognition sequences: ‘YVAD’ and ‘LEVD’, respectively) (Table 1). Furthermore, like caspases, they appear to be processive rather than degradative enzymes, as they are not active against a variety of general protease substrates nor do they exhibit any activity toward casine, bovine serum albumin or purified Rubisco. The latter is particularly worth noting as it reinforces that the function of the saspases in Rubisco cleavage is indirect rather than direct, likely by functioning in a protease cascade.
One major point of departure from caspases is the finding that upon induction of PCD by victorin, or heat shock, a treatment, which also induces PCD,42, 45, 46 saspase is detected in extracellular fluids by activity assays and inhibitor-labelled protein (biotin-YVAD-CMK). Importantly, saspase detection in the extracellular fluid is dependent on the induction of PCD. Although rapid secretion of the fully-processed form of the protein remains a possibility, because the total amount of labelled, processed saspase remains constant before and after PCD induction, yet upon induction of PCD becomes increasingly detectable in the extracellular fluid (despite treatment with cycloheximide), it appears likely that saspase is either relieved of inhibition or ‘untethered’ in the apoplast.
Phytaspases from Tobacco and Rice
A different approach to identifying plant analogues of caspases was initiated based on the finding that human caspase-3 is capable of inducing cleavage of the VirD2 protein encoded by the plant pathogenic bacterium, Agrobacterium tumefaciens. Because the VirD2 protein is known to operate inside Agrobacterium-infected plant cells to target insertion of a fragment of bacterial DNA (T DNA) into the plant cell's nuclear DNA, a search for a plant protease capable of cleaving VirD2 in a caspase-3-like manner (i.e., after the D400 residue within the TATD motif) was undertaken by Chichkova et al.47 Such a proteolytic activity was indeed detected in vivo using a VirD2-based reporter protein in tobacco (Nicotiana tabacum) leaves very soon (within 1 h) following induction of PCD by infection of N gene containing tobacco plants with tobacco mosaic virus (TMV). In this experimental model, the N gene product (an NB-LRR protein) mediates recognition of TMV, resulting in an HR, a form of plant PCD that limits virus spread. Tobacco plants that lack the N gene are unable to mount such a protective barrier and become infected owing to virus multiplication and spread throughout the plant. It is important to note that no caspase-3-like activity was detected in vivo in the absence of PCD: either in healthy leaves or in TMV-infected tobacco plants that lack the N gene.
In contrast to in planta studies showing that PCD induction is required for activation of the enzyme responsible for VirD2 fragmentation, active enzyme was found in leaf extracts of various plant species from both healthy and death-induced tissues.48 An explanation of this unanticipated phenomenon is that enzyme activation probably occurs during tissue wounding/disruption and/or upon incubation of extracts with the substrate protein. Thus, the enzyme responsible for VirD2 fragmentation was purified from healthy tobacco and rice (Oryza sativa) leaves and confirmed to possess caspase specificity in in vitro assays of peptide-based caspase substrates and inhibitors. It was able to hydrolyze a range of fluorogenic substrates, including AFC derivatives of VEID, VAD, YVAD, IETD, VDVAD and LEHD. For both tobacco and rice enzymes, VEID was the optimal recognition motif, whereas DEVD was completely inefficient (Table 1). Hence, the protease was named ‘phytaspase’, a plant aspartate-specific protease.49
Mass spectrometric characterization of tobacco and rice phytaspases revealed that both of them are subtilisin-like proteases. The cDNA sequences of both phytaspases were also identified and predicted to encode a 760-amino-acid-residue-long precursor protein, with an N-terminal signal peptide, followed by a prodomain (∼100 residues) and a protease domain (Figure 1). The canonical catalytic residues, Ser, Asp, and His, could be easily identified via sequence alignment with known subtilases of bacterial and plant origin. Tobacco and rice phytaspases share 53% amino-acid sequence identity.
Recombinant tobacco phytaspase was obtained by expressing the complete ORF in plants and was shown to possess the cleavage specificity of the native enzyme. Also like the native phytaspase, the mature recombinant enzyme lacked the signal peptide and prodomain. Mutation of the predicted catalytic Ser537 residue abolished both proteolytic activity and prodomain cleavage, indicating that proenzyme processing occurs autocatalytically. In accordance with the caspase-like cleavage specificity of phytaspase, the amino-acid residue preceding the prodomain cleavage site is D in both tobacco and rice phytaspases and mutation of this D residue impairs proenzyme processing.49
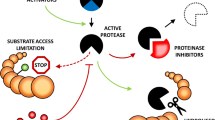
Phytaspase localization studies have revealed that, in healthy leaf tissues, the proenzyme is constitutively synthesized and processed, and the resultant mature enzyme lacking the prodomain is secreted from the cell into the apoplast (intercellular fluid)49 (Figure 2). This is in a sharp contrast to animal caspases that are intracellular enzymes. Within the apoplast, phytaspase may be sequestered before PCD and/or perhaps fulfils a defensive function, for example, by directly cleaving effectors secreted into the apoplast by pathogens. Notably, wounding/disrupting healthy leaf tissue produces an extract with a high level of phytaspase activity, suggesting that either the phytaspase is stored in an active form or that desequestration/activation occurs rapidly upon tissue disintegration.48
Processing and localization of phytaspase in healthy and dying plant cells, as compared with the behaviour of caspases in animal cells. Death proteases in animals (caspases) and in plants (phytaspases) are synthesized as inactive precursor proteins. Caspase precursors are stored within healthy animal cells until an apoptogenic stimulus triggers their processing/activation.50 In contrast to this scenario, phytaspase precursors are constitutively processed and the resultant mature enzymes secreted from healthy plant cells. Upon programmed cell death (PCD) induction, phytaspase is imported into plant cells through an undefined mechanism49
Surprisingly, upon induction of PCD by biotic (TMV infection) or abiotic (high salinity or oxidative stress) factors, phytaspase is relocalized from the apoplast into the cytoplasm, in which it is presumably capable of cleaving its intracellular targets (Figure 2). This is not a general default pathway induced by PCD, as other known (phytaspase-unrelated) proteases, such as cathepsin B, were shown to retain their apoplastic localization despite PCD induction.49 It appears that plants exploit a novel strategy to control their death protease(s): the enzyme is excluded (secreted) from healthy cells and allowed to enter the cell interior upon induction of PCD. In contrast to this scenario, animal caspases are controlled at the level of proenzyme processing/activation (Figure 2).50
Several lines of evidence show that phytaspase is an essential component of plant suicide machinery. In transgenic tobacco plants overexpressing phytaspase, PCD symptoms induced by biotic (TMV infection of N gene-carrying plants) and abiotic stresses (oxidative stress, osmotic stress) are markedly enhanced.49 Among such symptoms are: loss of cell viability, release of cytochrome c from mitochondria, accumulation of reactive oxygen species and induction of HSR203J, which is an early marker of the HR.49 Conversely, downregulation of phytaspase activity achieved either by direct enzyme inhibition or by siRNA-mediated silencing of phytaspase gene expression resulted in PCD suppression.49 The protective function of phytaspase-mediated HR is further supported by the observation that TMV accumulation in phytaspase-overproducing plants is markedly reduced, whereas in phytaspase-silenced plants it is enhanced, relative to control (wild-type) plants.49
Table 1 summarizes the current knowledge on phytaspases and saspases and enables their comparison with animal caspases.
Concluding Remarks and Future Perspectives
The current data indicate that the role played by caspases in animal PCD is taken, at least in part, by subtilisin-like proteases in plants. Despite the fact that caspases and subtilases are structurally completely unrelated, these proteases can display similar cleavage specificity and function. It will be interesting to decipher why the death proteases of both kingdoms appear to be D-specific proteases.
Subtilisin-like proteases of the S8A subfamily (which includes all plant subtilisin-like proteases) often lack stringent cleavage specificity.51, 52 Plants typically possesses dozens of subtilisin-like protease genes (over 50 members in Arabidopsis thaliana and over 60 members in O. sativa), but little evidence for exquisite substrate specificity of these proteolytic enzymes is available. Phytaspase and saspases with their ‘caspase-like’ specificity are clear exceptions to this rule. Phylogenetic comparison of phytaspases with the Arabidopsis subtilases represented by six distinct subgroups13 places phytaspases within subgroup 1 of the Arabidopsis subtilisin-like proteases and confirms the notion that phytaspases from tobacco and rice are close relatives (Figure 3). Furthermore, the putative subtilisin-like protease from rice (accession number BAB89803) that displays the highest similarity to sequenced peptides of the oat saspases also falls to the same subgroup and turns out to be highly related to rice phytaspase. Thus, aspartases appear to form a separate unit within the plant S8A subtilase subfamily. It would be of interest to learn how many subfamily members display Asp-specific proteolytic activity.
Phylogenetic comparison of phytaspases with Arabidopsis subtilisin-like proteases. Unrooted, neighbor-joining tree for Arabidopsis subtilases was generated as in Rautengarten et al.,13 with the addition of the Nicotiana tabacum and Oryza sativa phytaspase sequences (accession numbers GQ249168 and GI32977156, respectively) and of O. sativa putative ‘saspase-like’ BAB89803 sequence to the alignment
Although the ability of phytaspase and saspases to hydrolyze a number of peptide-based caspase substrates (Table 1) gives some illusion of enzyme promiscuity, comparative assays with several protein targets show that phytaspases possess even higher hydrolytic stringency than caspase-3 and therefore appear very selective.49 These are processive rather than degradative enzymes and as such are reminiscent of animal and yeast subtilisin-like proteases of S8B subfamily called pro-protein convertases. Convertases are involved in proteolytic processing of precursors to generate bioactive proteins and peptides.53 However, convertases introduce cleavage after a basic amino-acid residue (Lys, Arg), not after Asp.
Only a few proteases are known that display strict Asp cleavage specificity. To get an insight into phytaspase active-site architecture, we built a three-dimensional model of tobacco phytaspase based on the available structure of the SlSBT3 subtilase from tomato.15 Docking the Ac-VAD-CHO phytaspase inhibitor to the active site reveals several notable features (Figure 4). The inhibition-mediating active aldehyde group of Ac-VAD-CHO becomes settled in close proximity to the hydroxyl group of the catalytic Ser537, thus supporting the docking model. Interestingly, the S1 substrate-binding pocket accommodates the P1 Asp side chain in such a way that the β-carboxylic group is predicted (by DockingServer algorithms) to establish polar interactions with the imidazole ring of His331. The model thus suggests that His331 is involved in fixing the P1 Asp of the substrate, which provides perfect stereo conditions for the hydroxyl group of Ser537 to attack the peptide bond. In the rice phytaspase sequence (as well as in the putative ‘saspase-like’ BAB89803 sequence), the relevant position is also occupied by a His residue, and this particular His residue is absent from the majority of plant subtilases. Although the established theoretical model of the phytaspase active site requires experimental verification, it provides a rationale for the Asp specificity of the phytaspases and saspases. Of note, in caspases proper positioning of the P1 Asp is ensured by two Arg and one Gln residue.50
A model of the phytaspase active site. A homology model of processed tobacco phytaspase based on the structure of tomato SBT3 subtilase15 (PDB 3IS6) was built using MODELLER 9.7.56 Docking calculations for Ac-VAD-CHO phytaspase inhibitor were performed using DockingServer.57 The lowest free energy of binding model was visualized using PyMOL.58 Catalytic amino-acid residues Asp149, His220, Asn322 and Ser537 are shown in orange, with O atoms in red and N atoms in blue. The same designations are employed for His331 predicted to fix the β-carboxylic group of the ligand P1 Asp through polar interactions (circled). Ligand position (green) is shown from different angles in (a) and (b)
Proteolytic activities capable of hydrolyzing fluorogenic, peptide-based caspase substrates have been repeatedly observed in diverse plant PCD systems. Although phytaspase and saspases with their spectrum of recognized caspase substrates could account for the majority of caspase-like activities so far detected, there is at least one exception. Neither phytaspase nor saspase could hydrolyze a DEVD-based caspase substrate, yet ‘DEVDase’ activity has been frequently implicated in plant PCD models. It seems likely therefore that the family of plant ‘caspase-like’ proteases is currently incomplete. It is tempting to speculate that the DEVDase activity identified in oats as part of the same protease cascade involving saspase is also a saspase. If so, this ‘missing’ DEVDase activity could also be assigned to a subtilisin-like protease. However, another possibility has been recently shown for a DEVDase activity, the proteasome subunit, PBA1.9
The relationship between phytaspase and saspase is not completely clear at present. Both enzymes are subtilases, and display proteolytic activity towards a range of peptide-based caspase substrates with the notable exception of DEVD, and both enzymes can accumulate in the apoplast. Phytaspase activity, consistent with an essential role of this enzyme, is observed in a wide variety of plant species, including mono- and dicotyledonous plants.48 Saspases, while remaining uncharacterized at the genetic level, appear to be conserved at least in rice. Phytaspases from tobacco and rice, despite coming from two rather distant organisms, display similar substrate preferences (VEID-AFC is the optimal substrate, YVAD-, VAD-, IETD- and LEHD-AFC are somewhat less efficiently hydrolyzed, whereas WEHD-, VDVAD- and STATD-AFC are only marginally cleaved) (Table 1),49 whereas the saspases purified from oats, a close relative of rice, do not cleave either VEID, or WEHD or VDVAD derivatives at all and their optimal substrate is VKMD (Table 1).42 Whether these differences reflect different reaction conditions or there is a more principal distinction among the enzymes remains to be elucidated.
Redistribution of phytaspase from outside the cell (the apoplast) into the cytoplasm upon induction of PCD by biotic and abiotic stresses represents a novel phenomenon49 (Figure 2). Because inhibition of protein synthesis does not affect this process, cytoplasmic accumulation of phytaspase is not due to impaired secretion of the newly synthesized protein to the apoplast under PCD conditions. Rather, the enzyme appears to be physically and specifically relocalized from the apoplast to inside the cell. Although a mechanism underlying this phenomenon is not clear, a likely implication of this event is to give the death protease access to its intracellular targets only upon initiation of PCD, and to prevent unintended phytaspase-mediated proteolysis in the absence of death stimuli. This can be regarded as a plant-specific alternative to a well-known strategy employed by animals: to keep death proteases unprocessed (inactive) in the absence of death stimuli and to neutralize inappropriately activated caspases with specific protein inhibitors.
Upon entering the cell, phytaspase contributes to PCD accomplishment via hydrolysis of its intracellular target(s). Indeed, in phytaspase-silenced transgenic tobacco plants PCD is markedly suppressed, whereas cell death can be restored by expressing rice wild-type phytaspase, but not its proteolytically inactive mutant.49 Furthermore, phytaspase-mediated fragmentation of the intracellular reporter protein (VirD2-GFP) was shown to occur in response to death stimuli.47 It seems likely that cleavage of some phytaspase protein targets ultimately causes mitochondrial dysfunction, as exemplified by the correlation between the levels of phytaspase activity and cytochrome c release from mitochondria and H2O2 production in stressed transgenic plants.49
By analogy to the phytaspases, a similar ‘extracellular’ protective mechanism may be employed by the saspases. It is clear that at least one of the targets of saspase activation is the proteolytic processing of Rubisco, an intracellular target. However, saspase activity progresses from undetectable to high levels in the extracellular fluid upon induction of PCD. Although the mechanism regulating this change is unknown, given the rapidity of its appearance and the finding that the amount of total, processed enzyme does not increase nor is the extracellular increase blocked by cycloheximide, it likely arises due to de-tethering or unmasking of saspase activity in the extracellular space. Also, it is evident that the protease directly processing Rubisco is not an aspase, and that saspase acts through a protease cascade involving a DEVDase activity and a cysteine protease. Consequently, this extracellular activity must be transduced to the cytosol. To speculate based on the precedence set by the phytaspases, it is possible that some proportion of the saspase subsequently enters the cell upon activation, or that saspase activates the DEVDase activity (or some other enzyme) and the latter becomes internalized.
Knowledge of the protein targets for these Asp-specific subtilases is essential for unravelling molecular mechanisms of plant PCD. Thus far, the only known cellular substrates for phytaspase (as shown) and for the saspases (as inferred) are themselves. However, the characterization of Rubisco proteolysis during victorin and heat-shock-induced PCD led to the identification of at least two plant aspases, a DEVDase and the saspases that both appear to participate in a protease cascade leading to the degradation of their intracellular target. Furthermore, because heat shock is an abiotic stress and based on the recent findings that victorin-induced PCD is dependent on a gene encoding an NB-LRR protein, a type of protein that commonly regulates effector-triggered immunity, a role for saspases is implicated in both abiotic and biotic PCD responses.
For phytaspase, identification of the A. tumefaciens VirD2 protein as a death protease substrate had a profound impact on its identification and characterization. Phytaspase-mediated fragmentation of the VirD2 protein appears to represent a novel protective mechanism used by plants to limit transformation of plant cells by A. tumefaciens. Agrobacterium exploits VirD2 protein for the formation of a specific bacterial DNA strand (T DNA) and for its delivery into the nucleus of infected plant cells to achieve integration of the bacterial DNA into the plant genome (and plant transformation). For this purpose, VirD2 is equipped with a nuclear localization signal (NLS) positioned close to its C terminus.54 A single break introduced by phytaspase in this very region of VirD2 detaches the NLS and impairs nuclear import of VirD2-linked foreign DNA. In accord with this scenario, a mutant A. tumefaciens strain that encodes a phytaspase-resistant VirD2 protein instead of the wild-type one was shown to provide enhanced delivery and expression of foreign DNA in plants.55 Thus, in addition to its role in mediating plant PCD induced by abiotic stresses, phytaspase performs a protective role in response to biotic (pathogenic) insults, as exemplified by VirD2 fragmentation in the course of Agrobacterium infection, and by mediating the HR that limits TMV spread throughout the plant in the course of viral infection.
Besides the subtilisin-like proteases discussed above, several other plant proteases, such as metacaspases and VPE, also display similarities to animal caspases and are described in detail elsewhere in this CDD issue.
Accession codes
Abbreviations
- PCD:
-
programmed cell death
- HR:
-
hypersensitive response
- TMV:
-
tobacco mosaic virus
- NLS:
-
nuclear localization signal
References
Williams B, Dickman M . Plant programmed cell death: can’t live with it; can’t live without it. Mol Plant Pathol 2008; 9: 531–544.
Reape TJ, Molony EM, McCabe PF . Programmed cell death in plants: distinguishing between different modes. J Exp Bot 2008; 59: 435–444.
Reape TJ, McCabe PF . Apoptotic-like regulation of programmed cell death in plants. Apoptosis 2010; 15: 249–256.
del Pozo O, Lam E . Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr Biol 1998; 8: 1129–1132.
Bonneau L, Ge Y, Drury GE, Gallois P . What happened to plant caspases? J Exp Bot 2008; 59: 491–499.
del Pozo O, Lam E . Expression of the baculovirus p35 protein in tobacco affects cell death progression and compromises N gene-mediated disease resistance response to tobacco mosaic virus. Mol Plant Microbe Interact 2003; 16: 485–494.
Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M et al. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 2004; 305: 855–858.
Rojo E, Martín R, Carter C, Zouhar J, Pan S, Plotnikova J et al. VPEgamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr Biol 2004; 14: 1897–1906.
Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K et al. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev 2009; 23: 2496–2506.
Dodson G, Wlodawer A . Catalytic triads and their relatives. Trends Biochem Sci 1998; 23: 347–352.
Rawlings ND, Barrett AJ, Bateman A . MEROPS: the peptidase database. Nucleic Acids Res 2010; 38: D227–D233.
Tripathi LP, Sowdhamini R . Cross genome comparisons of serine proteases in Arabidopsis and rice. BMC Genomics 2006; 7: 200–231.
Rautengarten C, Steinhauser D, Büssis D, Stintzi A, Schaller A, Kopka J et al. Inferring hypotheses on functional relationships of genes: analysis of the Arabidopsis thaliana subtilase gene family. PLoS Comput Biol 2005; 1: e40.
Baker D, Sohl JL, Agard DA . A protein-folding reaction under kinetic control. Nature 1992; 356: 263–265.
Ottmann C, Rose R, Huttenlocher F, Cedzich A, Hauske P, Kaiser M et al. Structural basis for Ca2+-independence and activation by homodimerization of tomato subtilase 3. Proc Natl Acad Sci USA 2009; 106: 17223–17228.
Yamagata H, Masuzawa T, Nagaoka Y, Ohnishi T, Iwasaki T . Cucumisin, a serine protease from melon fruits, shares structural homology with subtilisin and is generated from a large precursor. J Biol Chem 1994; 269: 32725–32731.
Schaller A . A cut above the rest: the regulatory function of plant proteases. Planta 2004; 220: 183–197.
Von Groll U, Berger D, Altman T . The Subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 2002; 14: 1527–1539.
Tanaka H, Onouchi H, Kondo M, Hara-Nishimura I, Nishimura M, Machida C et al. A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development 2001; 128: 4681–4689.
Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Büssis D, Altmann T . A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J 2008; 54: 466–480.
Tan-Wilson AL, Liu X, Chen R, Qi X, Wilson KA . An acidic amino acid-specific protease from germinating soybeans. Phytochemistry 1996; 42: 313–319.
Boyd PM, Barnaby N, Tan-Wilson A, Wilson KA . Cleavage specificity of the subtilisin-like protease C1 from soybean. Biochim Biophys Acta 2002; 1596: 269–282.
Liu JX, Srivastava R, Che P, Howell SH . Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J 2007; 51: 897–909.
Wolf S, Rausch T, Greiner S . The N-terminal pro region mediates retention of unprocessed type-I PME in the Golgi apparatus. Plant J 2009; 58: 361–375.
Srivastava R, Liu JX, Guo H, Yin Y, Howell SH . Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J 2009; 59: 930–939.
Srivastava R, Liu JX, Howell SH . Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J 2008; 56: 219–227.
Jordá L, Coego A, Conejero V, Vera P . A genomic cluster containing four differentially regulated subtilisin-like processing protease genes is in tomato plants. J Biol Chem 1999; 274: 2360–2365.
Tornero P, Conejero V, Vera P . Primary structure and expression of a pathogen-induced protease (PR-P69) in tomato plants: similarity of functional domains to subtilisin-like endoproteases. Proc Natl Acad Sci USA 1996; 93: 6332–6337.
Wolpert TJ, Macko V, Acklin W, Jaun B, Seibl J, Meili J et al. Structure of victorin C, the major host-selective toxin from Cochliobolus victoriae. Experientia 1985; 41: 1524–1529.
Wolpert TJ, Macko V, Acklin W, Jaun B, Arigoni D . Structure of minor host-selective toxins from Cochliobolus victoriae. Experientia 1986; 42: 1296–1299.
Wolpert TJ, Dunkle LD, Ciuffetti LM . Host-selective toxins and avirulence determinants: What's in a name? Ann Rev Phytopathol 2002; 40: 251–285.
Welsh JN, Peturson B, Machacek JE . Associated inheritance of reaction to races of crown rust, Puccinia coronata avenae Erikss., and to Victoria blight, Helminthosporium victoriae Mand M., in oats. Can J Bot 1954; 32: 55–68.
Mayama S, Bordin APA, Morikawa T, Tanpo H, Kato H . Association of avenalumin accumulation with co-segregation of victorin sensitivity and crown rust resistance in oat lines carrying the Pc-2 gene. Physiol Mol Plant Pathol 1995; 46: 263–274.
Lorang JM, Carkaci-Salli N, Wolpert TJ . Identification and characterization of victorin sensitivity in Arabidopsis thaliana. Mol Plant Microb Interact 2004; 17: 577–582.
Lorang JM, Sweat TA, Wolpert TJ . Plant disease susceptibility conferred by a ‘resistance’ gene. Proc Natl Acad Sci USA 2007; 37: 14861–14866.
Sweat TA, Lorang JM, Bakker EG, Wolpert TJ . Characterization of natural and induced variation in the LOV1 gene, a CC-NB-LRR gene conferring victorin sensitivity and disease susceptibility in Arabidopsis. Mol Plant Microbe Interact 2008; 21: 7–19.
Yao N, Tada Y, Park P, Nakayashiki H, Tosa Y, Mayama S . Novel evidence for apoptotic cell response and differential signals in chromatin condensation and DNA cleavage in victorin-treated oats. Plant J 2001; 28: 13–26.
Curtis MJ, Wolpert TJ . The victorin induced mitochondrial permeability transition precedes cell shrinkage and biochemical markers of cell death and shrinkage occurs without loss of membrane integrity. Plant J 2004; 38: 244–259.
Navarre DA, Wolpert TJ . Victorin induction of an apoptotic/senescence-like response in oats. Plant Cell 1999; 11: 237–249.
Tada Y, Hata S, Takata Y, Nakayashiki H, Tosa Y, Mayama S . Induction and signaling of an apoptotic response typified by DNA laddering in the defense response of oats to infection and elicitors. Mol Plant Microbe Interact 2001; 14: 477–486.
Curtis MJ, Wolpert TJ . The oat mitochondrial permeability transition and its implication in victorin binding and induced cell death. Plant J 2002; 29: 295–312.
Coffeen WC, Wolpert TJ . Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in oats. Plant Cell 2004; 16: 857–873.
Powers JC, Asgian JL, Ekici OD, James KB . Irreversible inhibitors of serine cysteine and threonine proteases. Chem Rev 2002; 102: 4639–4750.
Bernard D, Méhul B, Thomas-Collignon A, Delattre C, Donovan M, Schmidt R . Identification and characterization of a novel retroviral-like aspartic protease specifically expressed in human epidermis. J Invest Dermatol 2005; 125: 278–287.
Balk J, Leaver CJ, McCabe PF . Translocation of cytochrome c from the mitochondria to the cytosol occurs during heat-induced programmed cell death in cucumber plants. FEBS Lett 1999; 463: 151–154.
Tian R-H, Zhang G-Y, Yan C-H, Dai Y-R . Involvement of poly(ADP-ribose) polymerase and activation of caspase-3-like protease in heat shock-induced apoptosis in tobacco suspension cells. FEBS Lett 2000; 474: 11–15.
Chichkova NV, Kim SH, Titova ES, Kalkum M, Morozov VS, Rubtsov YP et al. A plant caspase-like protease activated during the hypersensitive response. Plant Cell 2004; 16: 157–171.
Chichkova NV, Galiullina RA, Taliansky ME, Vartapetian AB . Tissue disruption activates a plant caspase-like protease with TATD cleavage specificity. Plant Stress 2008; 2: 89–95.
Chichkova NV, Shaw J, Galiullina RA, Drury GE, Tuzhikov AI, Kim SH et al. Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. EMBO J 2010; 29: 1149–1161.
Fuentes-Prior P, Salvesen GS . The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J 2004; 384: 201–232.
Cedzich A, Huttenlocher F, Kuhn BM, Pfannstiel J, Gabler L, Stintzi A et al. The protease-associated domain and C-terminal extension are required for zymogen processing, sorting within the secretory pathway, and activity of tomato subtilase 3 (SlSBT3). J Biol Chem 2009; 284: 14068–14078.
Fontanini D, Jones BL . SEP-1-a subtilisin-like serine endopeptidase from germinated seeds of Hordeum vulgare L.cv. Morex. Planta 2002; 215: 885–893.
Steiner DF . The proprotein convertases. Curr Opin Chem Biol 1998; 2: 31–39.
Howard EA, Zupan JR, Citovsky V, Zambryski PC . The VirD2 protein of A. tumefaciens contains a C-terminal bipartite nuclear localization signal: implications for nuclear uptake of DNA in plant cells. Cell 1992; 68: 109–118.
Reavy B, Bagirova S, Chichkova NV, Fedoseeva SV, Kim SH, Vartapetian AB et al. Caspase-resistant VirD2 protein provides enhanced gene delivery and expression in plants. Plant Cell Rep 2007; 26: 1215–1219.
Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinform 2006; Chapter 5, Unit 5.6.
Bikadi Z, Hazai E . Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J Cheminform 2009; 1: 15.
Ordog R . PyDeT, a PyMOL plug-in for visualizing geometric concepts around proteins. Bioinformation 2008; 2: 346–347.
Acknowledgements
Parts of this work were supported by the Russian Foundation for Basic Research (NVCh and ABV) and by Russian Ministry of Education and Science (contracts P334 and 14.740.11.0168 (ABV, NVCh and AIT); contract number 02.740.11.5145 (MT and NVCh)), Royal Society (MT and ABV) and Scottish Government (MT). Work in the Wolpert lab is currently supported by grants from the United States Department of Agriculture National Research Initiative, Cooperative State Research, Education and Extension Service, Grant No. 2007-01598 and from the National Science Foundation Grant No. IOS-0724954.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by P Bozhkov
Rights and permissions
About this article
Cite this article
Vartapetian, A., Tuzhikov, A., Chichkova, N. et al. A plant alternative to animal caspases: subtilisin-like proteases. Cell Death Differ 18, 1289–1297 (2011). https://doi.org/10.1038/cdd.2011.49
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2011.49
Keywords
This article is cited by
-
XCP1 cleaves Pathogenesis-related protein 1 into CAPE9 for systemic immunity in Arabidopsis
Nature Communications (2023)
-
Genome-Wide Identification and Characterization of the SBT Gene Family in maize and Its Expression in the Various tissues
Plant Molecular Biology Reporter (2023)
-
Plant cell responses to allelopathy: from oxidative stress to programmed cell death
Protoplasma (2022)
-
Phytohormone treatment induces generation of cryptic peptides with antimicrobial activity in the Moss Physcomitrella patens
BMC Plant Biology (2019)
-
Phaeobacter inhibens induces apoptosis-like programmed cell death in calcifying Emiliania huxleyi
Scientific Reports (2019)