Abstract
Caspase-8 stably inserts into the mitochondrial outer membrane during extrinsic apoptosis. Inhibition of caspase-8 enrichment on the mitochondria impairs caspase-8 activation and prevents apoptosis. However, the function of active caspase-8 on the mitochondrial membrane remains unknown. In this study, we have identified a native complex containing caspase-8 and BID on the mitochondrial membrane, and showed that death receptor activation by Fas or tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induced the cleavage of BID (tBID formation) within this complex. tBID then shifted to separate mitochondria-associated complexes that contained other BCL-2 family members, such as BAK and BCL-XL. We report that cells stabilize active caspase-8 on the mitochondria in order to specifically target mitochondria-associated BID, and that BID cleavage on the mitochondria is essential for caspase-8-induced cytochrome c release. Our findings indicate that during extrinsic apoptosis, caspase-8 can specifically target BID where it is mostly needed, on the surface of mitochondria.
Similar content being viewed by others
Main
Programmed cell death or apoptosis is characterized by the activation of a class of proteases called caspases. Caspases are cysteine-dependent aspartic acid-specific proteases, which represent the executioners of apoptosis and are generally classified as either initiators or effectors. In mammalian cells, caspases are almost universally expressed as inactive zymogens, which must undergo proteolysis at an internal aspartate to produce the active form. Depending on the death-inducing signal, caspase activation and apoptosis occur via two distinct pathways: intrinsic or extrinsic. Intrinsic apoptosis is triggered by a number of different cellular stresses that cause the release of apoptogenic factors from the mitochondrial intermembrane space into the cytosol.1, 2 Mitochondrial outer membrane permeabilization (MOMP) is regulated by the interplay between the BCL-2 family of proteins and requires BAX and/or BAK activation.3, 4 In contrast to intrinsic apoptosis, extrinsic apoptosis is initiated at the plasma membrane by stimulation of death receptors.5 Binding of a death ligand, such as Fas or tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), triggers receptor oligomerization and induces the binding of adapter molecules.6 These adapter molecules recruit caspase-8 to the receptor complex to form the death-inducing signalling complex (DISC). Caspase-8 typically exists as a monomer in the cytosol, but upon DISC formation, it undergoes dimerization.7, 8 The dimerization of two caspase-8 monomers (p55/p55) results in the conformational change, which exposes the active site of the caspase through the so-called mechanism of ‘induced proximity’.7, 9, 10, 11 Dimerization has been demonstrated to be sufficient for activation of caspase-8, but it has been proposed that full activity requires self-cleavage.11, 12, 13, 14 Caspase-8 initially cleaves itself between the p18 and p10 domains forming a heterodimer within a heterotetrameric complex (p43-p10/p43-p10) (See Figure 1a). This first cleavage is necessary for recognition of other substrates, such as effector caspases (i.e. caspase-3) and the proapoptotic BCL-2 family member BID.12, 13 p43-p10/p43-p10 can undergo a second cleavage between the p26 and p18 domains, which frees the heterotetramer (p18-p10/p18-p10) from the DED domains but does reduce its activity.12
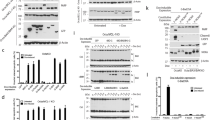
Analysis of the autoprocessing of exogenously expressed caspase-8-GFP mutants in shCaspase-8 HeLa cells. (a) Linear scheme depicting the location and molecular masses of the three functional domains of caspase-8. The sites of the first and second cleavages are labeled. p26 contains the DED domains and p43 is made up of the DED domains and p18 domain. The amino-acid substitutions of the caspase-8-GFP mutants are indicated in the table. (b) Immunoblot analysis of whole-cell lysates from shCaspase-8 HeLa cells transfected with the caspase-8-GFP point mutants. Fas-treated cells were incubated in 0.5 μg/ml anti-Fas for 14 h. (c) shCont (black bars) and shCaspase-8 (gray bars) HeLa cells were transfected with the various caspase-8-GFP mutants and assayed for cell death by flow cytometry. Only GFP-positive cells were counted and cell death was measured by PI staining. Fas conditions were the same as in (a). Error bars represent S.D. of three or more experiments done in triplicate
Extrinsic apoptosis can proceed along two different pathways, type I or type II, depending on the level of caspase-8 activation upon DISC formation.15 In type I cells, large amounts of DISC and active caspase-8 are formed that leads to the direct cleavage of effector caspases in the cytosol.16 In type II cells, there is slower DISC assembly and low production of active caspase-8.15 In this scenario, caspase-8 must engage the intrinsic (mitochondrial) pathway to amplify the death signal and execute apoptosis. The transition from the extrinsic pathway to the intrinsic pathway is achieved through caspase-8 processing of BID.17, 18 Cleaved/truncated BID, or tBID, interacts with other BCL-2 family members on the surface of the mitochondria, which results in MOMP.
We had previously demonstrated that during type II extrinsic apoptosis caspase-8 becomes enriched on the mitochondrial surface, and that by blocking caspase-8 association with the mitochondria, we were able to inhibit the formation of p43-p10, prevent BID cleavage and block apoptosis.19, 20 However, the purpose of the insertion of active caspase-8 into the mitochondrial membrane remains unknown. In this study, we report that caspase-8 and BID form a native macromolecular complex on the mitochondria. Following Fas or TRAIL activation, tBID is formed within this complex by the caspase-8-mediated cleavage of BID and then shifts to a separate complex containing BAK and/or BCL-XL. By concentrating BID and caspase-8 on the mitochondrial membrane, type II cells have established a mechanism by which low levels of proteolytically active caspase-8 would be able to specifically target sufficient amounts of BID on the organelle (mitochondria) where it is needed to induce MOMP. These findings further emphasize the important role of compartmentalization to the activation of caspases, such as is seen with the apoptosome and inflammasome.21, 22 In summary, we have identified a mitochondria-associated triggering platform from which type II cells launch the mitochondrial phase of apoptosis.
Results
The generation of caspase-8 point mutants to study mitochondrial-dependent cell death
We hypothesized that point mutations that affect caspase-8 processing and activity might also influence the association of caspase-8 with the mitochondria. Figure 1a depicts a linear cartoon of caspase-8 showing the main functional domains. Caspase-8 is cleaved into three distinct fragments (DED, p18 and p10) whose boundaries and molecular weights are labeled in Figure 1a. Five different mutant constructs were created that specifically targeted aspartate residues in the regions of the first and second cleavage sites of caspase-8 or the active cysteine (Figure 1a). All constructs were tagged with GFP in order to examine localization and to distinguish exogenous from endogenous caspase-8 (Figure 1a).
We initially explored the cleavage patterns of the caspase-8-GFP point mutants. For that reason, HeLa cells, which stably expressed an shRNA against endogenous caspase-8 (Supplementary Figure S1), were transfected with the different caspase-8-GFP mutants. The formation of both p43 and p18 was evident in cells transfected with wt caspase-8 (C8-GFP), or with caspase-8 mutated at the second cleavage site (DM2-GFP or TM2-GFP), but not in cells transfected with caspase-8 mutated at the first cleavage site (Figure 1b). Furthermore, the inactive caspase-8 mutant, C360S-GFP, displayed a strong inhibition of p43 and p18 formation, suggesting a defect in self-cleavage (Figure 1b, lane 8).
Given the observed alterations in the cleavage pattern of the caspase-8 point mutants, we next sought to analyze their effects on the induction of cell death. Surprisingly, all caspase-8 mutants were capable of inducing cell death in control HeLa cells expressing a non-targeting shRNA (shCont) to a level similar to Fas treatment (Figure 1c, black bars), despite the defects in autoprocessing noted in Figure 1b. It should be noted that, in the absence of death ligand, exogenous expression of caspase-8 induces its dimerization through a process termed ‘induced-proximity’.7, 10, 13 Importantly, caspase-8 was not highly overexpressed, and most transfected cells displayed a ∼2-fold increase in the total amount of caspase-8 (Supplementary Figure S2a). It is likely that dimerization occurred between exogenously expressed inactive caspase-8 mutants (DM1-GFP and C360S-GFP) and endogenous caspase-8. Indeed, the second cleavage product of caspase-8, p18, was observed in shCont cells transfected with the different active and inactive caspase-8 mutants, but was absent in shCaspase-8 cells transfected with the inactive forms of caspase-8 (Supplementary Figure S2b). Of note, less p18 formation was observed in control cells transfected with DM1-GFP, which cannot undergo the first cleavage on its own (Supplementary Figure S2b). This is because of the fact that, unlike all the other caspase-8 constructs, the p18 formation in cells transfected with this mutant is fully dependent on the cleavage of endogenous caspase-8. Next, the effect of the different caspase-8 mutants on viability was assessed in shCaspase-8 HeLa cells. Indeed, these cells were protected from Fas-induced apoptosis (Figure 1c, gray bars). However, the expression of C8-GFP, DM2-GFP, TM2-GFP or SM1-GFP caused levels of cell death similar to that in shCont HeLa, indicating that these exogenously expressed caspase-8 constructs are fully capable of inducing apoptosis. In contrast, the expression of DM1-GFP or C360S-GFP did not induce cell death without the presence of endogenous caspase-8 (Figure 1c).
Mitochondrial localization of caspase-8 point mutants
In order to evaluate the link between caspase-8 processing and the subcellular localization, shCont HeLa cells were transfected with the different caspase-8-GFP mutants and assessed by confocal microscopy using the mitochondria-tagged protein Mito-dsRed2 as a marker. Exogenously expressed wt caspase-8 was mostly observed on the mitochondria (Figure 2). Furthermore, we confirmed that the mitochondrial localization of caspase-8 occurred before cytochrome c release and was not blocked by BCL-XL overexpression (Supplementary Figure 3). The disruption of the second cleavage site of caspase-8 still resulted in mitochondrial localization (Figure 2). In addition, non-mitochondrial punctated structures were consistently observed (Figure 2, white arrows). In contrast, when the first cleavage site of caspase-8 was disrupted, the protein lost its mitochondrial localization and was then diffused in the cytosol (Figure 2). This suggested that the formation of the p43-p10 heterodimer may be needed for stable enrichment of caspase-8 on the mitochondria. It is important to mention that even when exogenously expressed caspase-8 was present on the mitochondria, a significant amount of it was still in its unprocessed form (Figures 1b and 3a–c). The inactive C360S caspase-8 mutant was also observed on the mitochondria (Figure 2). This is in line with the fact that unlike DM1-GFP, this mutant is capable of being processed into the p43-p10 form by the endogenous caspase-8 through intra- or interdimer cleavage.13 Indeed, C360S-GFP did interact with endogenous caspase-8 on the mitochondria (Figure 3a), and can produce high amounts of mitochondria-associated p43 in shCont HeLa (Figure 3b). The association of the p43 form of C360S-GFP with the mitochondria was completely absent in shCaspase-8 cells, demonstrating the need for the endogenous caspase-8 for the first cleavage of this mutant (Figure 3c). Furthermore, unlike the active mutant TM2-GFP, the two inactive mutants DM1-GFP and C360S-GFP could not generate mitochondria-associated caspase-8-like activity (LETDase) without endogenous caspase-8 (Figure 3d). Overall, these results are in line with the observed dependency of these inactive mutants on endogenous caspase-8 for cell death induction (Figure 1c).
Caspase-8-GFP translocation to the mitochondria is dependent on the formation of p43-p10. Confocal images of shCont HeLa transfected with caspase-8-GFP mutants. z-VAD-fmk, a cell-permeable pancaspase inhibitor, was added before transfection and kept throughout the experiment to prevent cell death. Expression of Mito-dsRed2 was used as a mitochondrial marker. White bars=10 μm. White arrows mark the presence of punctated non-mitochondrial caspase-8-GFP
p43 formation is necessary for translocation and activity of caspase-8 on the mitochondria. (a) Mitochondria-enriched fractions were generated from C360S-GFP or pEGFP-N1-transfected shCont HeLa cells. Mitochondrial lysates were pelleted and the supernatant was then immunoprecipitated with anti-GFP and immunoblotted for caspase-8, GFP and VDAC1. (b and c) Mitochondria were isolated from pEGFP-N1, TM2-GFP, DM1-GFP and C360S-GFP shCont or shCaspase-8 HeLa cells. Blots were probed with anti-cleaved caspase-8 (D374), anti-GFP and anti-TOM20. Note the lack of p10-GFP formation and low levels of p43 on DM1-GFP mitochondria. All lanes in (c) were taken from the same exposure. (d) The mitochondria used in (c) were used to assess the activity of mitochondria-bound caspase-8. The level of caspase-8-like activity (zLETD-aminoluciferin cleavage) is displayed relative to the GFP control (defined as zero) in units of luminescence (RLU). Note the similar levels of caspase-8 activity in DM1-GFP and C360S-GFP cells. Error bars represent S.D. (n=3)
BID and caspase-8 form a native complex on the mitochondrial membrane
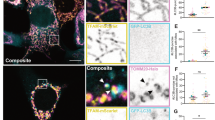
Our data so far has suggested that the mitochondrial localization of active caspase-8 may be needed to specifically target a protein(s) on the mitochondria to execute apoptosis. As it was previously shown that type II extrinsic apoptosis is dependent on BID cleavage by caspase-8, we sought to determine if caspase-8 forms a complex with BID on the mitochondria. Mitochondria isolated from shCont HeLa cells, which had been transfected with either BID-mCherry or C360S-mCherry, were solubilized in the mild non-ionic detergent, dodecyl-β-D-maltoside (DDM), and the lysates (supernatant) were run by clear native electrophoresis (CNE). Figure 4a clearly showed that both BID-mCherry and C360S-mCherry formed a >200 kDa complex, which co-migrated to the same position in the first dimension. The BID-mCherry and C360S-mCherry lanes were excised and run in the second dimension by SDS-PAGE to dissociate the complex. Immunoblotting indicated that the majority of mitochondria-associated endogenous BID (and BID-mCherry) was present within the complex (Figure 4b, dashed line). Furthermore, 2D CNE/SDS-PAGE revealed that endogenous caspase-8 co-migrated with BID-mCherry, suggesting that it was part of the native complex. Identical results were obtained from the reciprocal experiment using C360S-mCherry mitochondria, which showed that endogenous BID co-migrated with both C360S-mCherry and endogenous caspase-8 (Figure 4c, dashed line). This was the first evidence of caspase-8 and BID forming a native complex on the mitochondrial membrane. Importantly, active caspase-8 (p43) was observed within this large complex in caspase-8-transfected cells (Figure 4d and Supplementary Figure S4). The mitochondria-associated BCL-2 family proteins, BCL-XL and BAK, were not part of the caspase-8/BID complex (Figure 4b and c).
Caspase-8 and BID form a high-molecular weight complex on the mitochondrial membrane. (a) shCont HeLa were transfected with BID-mCherry or C360S-mCherry. 50 μM zVAD-fmk was added to C360S-mCherry-transfected cells to prevent cell death. Isolated mitochondria were solubilized in DDM and run on a 4–16% native gel. The gel was scanned for direct comparison of fluorescent signals. (b and c) Lanes from the gel in (a) were excised, denatured and run in the second dimension by SDS-PAGE. Membranes were immunoblotted for caspase-8 and the BCL-2 family proteins BID, BAK, BCL-XL. Complex III was used as a control. (d) Mitochondria isolated from shCont HeLa transfected with TM2-GFP were treated as in (a). The presence of p43 was clearly detected on the 200 μg immunoblot. In all, 100 μg of mitochondria was used for all subsequent 2D CNE/SDS-PAGE experiments for clarity purposes
Fas- or TRAIL-activation induced cleavage of BID within a native complex on the mitochondrial membrane
Having identified a native complex of caspase-8 and BID, we wanted to examine the effects that activation of caspase-8 had on the state of the complex. Initially, we found that the endogenous caspase-8/BID complex was also resident on the mitochondrial membranes of untransfected and untreated HeLa cells (Figure 5a, solid line). This was supported by 1D SDS-PAGE analysis of isolated mitochondria, which confirmed the presence of BID and caspase-8 on naïve mitochondria (Figure 5c, lanes 1–2). The localization of full length BID on mitochondria of untreated cells would be somewhat surprising, as BID has been reported to be a cytosolic factor that translocates to the mitochondria only after cleavage by caspase-8.17, 18 Nevertheless, we consistently detected BID on the mitochondria, and after subcellular fractionation analysis estimated that ∼20% of full-length BID protein was associated with the mitochondria of untreated cells (data not shown). We also investigated if this complex was present in the non-small-cell lung carcinoma cell line H460. H460 are type II cells and are sensitive to TRAIL-induced extrinsic apoptosis.23 Similar to HeLa cells, untreated H460 cells contained a native caspase-8/BID complex on the mitochondria (Figure 6a). Taken together, these results indicate that both BID (full-length) and procaspase-8 (p55) naturally reside on the mitochondria within a large molecular weight complex.
Fas activation induces the cleavage of BID within the caspase-8/BID complex. (a) Mitochondria from untreated shCont HeLa were subjected to 2D CNE/SDS-PAGE. Caspase-8 and BID form a native complex on the mitochondrial membrane (solid line). (b) Fas activation led to appearance of tBID in the caspase-8/BID complex (solid line). tBID also comigrated with BAK and/or BCL-XL, suggesting the formation of other complexes on the mitochondrial membrane (dotted lines). A plot profile was generated from the immunoblots shown in the top panel using the software program, ImageJ (http://rsb.info.nih.gov/ij/). Note that the majority of the caspase-8, BID and tBID on the mitochondrial membrane is within a single complex. Additionally, there were large decreases in the levels of full-length BID on the mitochondrial membrane after Fas treatment (compare BID panel of (a) with that of (b)). (c) DDM-resistant fractions from (a) and (b) were analyzed by SDS-PAGE after solubilizing the pellet in 1% SDS. p43, and a substantial proportion of p55 and BID, were preferentially retained in the pellet compared with the other mitochondrial proteins examined. Again, there were large decreases in the levels of full-length BID on the mitochondrial membrane after Fas treatment (compare lanes 1–2 with 3–4). Lower panel; the LETDase activity of the DDM-soluble fractions from untreated or αFas-treated shCont HeLa mitochondria was measured as described in Materials and methods. A substantial increase in caspase-8 activity was seen in the DDM-soluble fraction from αFas-treated compared with untreated cells
TRAIL activation induces the cleavage of BID within the caspase-8/BID complex. (a and b) Mitochondria from untreated H460 cells were subjected to 2D CNE/SDS-PAGE. As seen in HeLa cells, caspase-8 and BID form a native complex on the mitochondrial membrane (solid line). (b) TRAIL activation led to the appearance of tBID in the caspase-8/BID complex (solid line). tBID also comigrated with BAK and/or BCL-XL (dotted lines). (c) The DDM-resistant fraction from the mitochondria was analyzed for the presence of active (p43) caspase-8. Similar to HeLa cells, a substantial proportion of p55 and p43 were present in the detergent-resistant fraction. Also, note that tBID was detectable in the soluble fraction. (Lower panel) The LETDase activity of the DDM-soluble fractions from untreated or TRAIL-treated H460 mitochondria was measured as in Figure 5c. A substantial increase in caspase-8 activity was seen in the DDM-soluble fraction from TRAIL-treated compared with untreated cells. (d) Subcellular fractionation of H460 cells±TRAIL revealed that p43 is specifically enriched on the mitochondria of H460 cells after TRAIL activation (compare lanes 3 and 4), and that BID on the mitochondria was preferentially processed by caspase-8 in comparison with cytosolic BID
Marked differences in the constitution and migration of the mitochondrial complexes were observed when caspase-8 was activated by TM2-GFP transfection or death receptor ligation (Figures 4d, 5b and 6b). The initiation of apoptosis led to an increase in the amount of activated caspase-8 (p43) on the mitochondria (Figure 6d, compare lanes 3 and 4). Although p43 was detectable in the DDM-soluble fraction of the mitochondria, it was found to be more highly enriched in the DDM-resistant fraction (Figure 5c, compare lanes 3 and 4). In contrast, BID and procaspase-8 were more equally distributed between the fractions, whereas tBID, BAK, BCL-XL and VDAC1 were more soluble (Figures 5c and 6c). Figure 4d clearly showed that in TM2-GFP-transfected cells, p43 was also present in the caspase-8/BID complex. Similar results were obtained in C360S-GFP-transfected HeLa cells (Supplementary Figure S4). In agreement, there was a substantial increase in caspase-8 LETDase activity in the DDM-soluble fractions of Fas-treated shCont HeLa and TRAIL-treated H460 cells compared with untreated cells (Figures 5c and 6c, lower panel). In parallel with p43 formation, we found that tBID was also formed within the native complex with caspase-8 (Figures 5b and 6b, solid line) and shifted to larger molecular weight complexes, which co-migrated with both BAK and BCL-XL (dotted lines). These results suggested that after cleavage of BID within the caspase-8/BID complex, tBID is potentially targeted to a separate set of complexes with the BCL-2 family members BAK and/or BCL-XL. A plot profile generated from densitometric analysis of the immunoblots in Figure 5b showed that the majority of the various mitochondria-bound forms of caspase-8 and BID were within a single complex on the mitochondria (Figure 5b, lower panel).
Recombinant caspase-8 is sufficient to induce cytochrome c release from isolated mitochondria
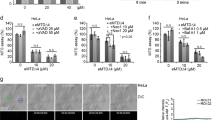
To further characterize the activity of caspase-8 on mitochondria recombinant, active caspase-8 protein (rCaspase-8) was used. Initially, we tested whether active caspase-8 was sufficient to cause the permeabilization of isolated mitochondria. The level of caspase-8 activity on mitochondrial membranes after incubation with rCaspase-8 was comparable with mitochondria isolated from Fas-treated HeLa cells as measured by an LETDase activity assay (Supplementary Figure S5). rCaspase-8 caused a dose-dependent release of cytochrome c from mitochondria isolated from HeLa cells (Figure 7a). This was the first indication that incubation of isolated mitochondria with active caspase-8 is sufficient for MOMP. As MOMP is regulated by the BCL-2 family of proteins,24 we assessed whether the release of cytochrome c induced by rCaspase-8 could be blocked by BCL-XL. Incubation of mitochondria isolated from BCL-XL-expressing HeLa cells with rCaspase-8 showed that cytochrome c release was blocked (Figure 7b, compare lanes 2 and 5). These results suggested that caspase-8-induced cytochrome c release from isolated mitochondria was dependent on the BCL-2 family of proteins.
Caspase-8 specifically targets BID on the mitochondrial membrane to induce MOMP. (a and b) Mitochondria isolated from shCont HeLa (or BCL-XL HeLa) by subcellular fractionation were incubated with different concentrations of recombinant active caspase-8 (rC8). Mitochondria were separated from the supernatant by centrifugation, and both fractions were immunoblotted for cytochrome c release. The Triton X-100 lane represents the total amount of cytochrome c in the purified mitochondria (c) Mitochondria were isolated from shCont cells transfected with a non-targeting siRNA (siCont) or an siRNA against BID (siBID). Mitochondria were treated as described in (a)
BID is necessary for caspase-8-induced cytochrome c release from isolated mitochondria
Figure 7b suggested that caspase-8-induced cytochrome c release is regulated by BCL-2 proapoptotic proteins. BID is known to be a substrate of caspase-8 after death receptor activation,25, 26 and we identified a native mitochondrial complex containing caspase-8 and BID (Figures 4,5,6). Therefore, we examined if caspase-8 could cleave mitochondria-associated BID and if this was sufficient to induce MOMP. Indeed, tBID formation was observed in mitochondria treated with rCaspase-8 (Figure 7c, lane 2). Although mitochondria-associated BID was cleaved by caspase-8, there may be other targets of caspase-8 on the mitochondrial membrane, which are necessary for MOMP. To test the dependence of caspase-8 on BID to induce MOMP, we transfected HeLa cells with either a non-targeting control siRNA (siCont) or an siRNA against BID (siBID) before mitochondria isolation. rCaspase-8 induced the release of the majority of cytochrome c from the mitochondria isolated from siCont cells (Figure 7c, lane 2). In contrast, mitochondria isolated from siBID-transfected cells displayed a significant reduction in the amount of cytochrome c release and tBID formation (Figure 7c, lane 4). These results indicated that BID was required for rCaspase-8-induced cytochrome c release, and that the amount of BID present on the mitochondrial membranes is sufficient to support MOMP. In addition, Figure 7c indicated that caspase-8 association with the mitochondria is not dependent on the availability of BID on the mitochondrial membrane (compare lanes 2 and 4). In addition, we found that TRAIL activation of H460 cells resulted in significant decreases in the levels of mitochondria-bound BID as compared with cytosolic BID, suggesting that the mitochondria may be the main site of caspase-8-induced cleavage of BID (Figure 6d).
Discussion
Numerous studies have reported difficulty in detecting active caspase-8 (p43-p10) at the DISC in type II cells.15, 27 We have now shown that, after Fas or TRAIL activation, p43-p10 is readily detectable on the mitochondrial membrane in multiple cells lines. Given that a native caspase-8 population is already present on the mitochondrial membrane, it is tempting to speculate that caspase-8 can undergo dimerization and self-cleavage/activation directly upon the mitochondrial membrane. Alternatively, the presence of non-mitochondrial punctated structures (Figure 2, white arrows) could suggest that p43-p10 is formed at the DISC and subsequently translocates to the mitochondria. Mitochondrial targeting of active caspase-8 may be needed for the activation of the native caspase-8/BID complexes on the outer membrane. If this scenario were true, one would expect to see p43-p10 enrichment on the mitochondrial membranes of type I cells. Indeed, we did observe tBID formation within the caspase-8/BID complex in cells, which underwent a ‘type-I-like’ mitochondrial-independent apoptosis (Supplementary Figure S6). We are currently investigating the mechanism and location of caspase-8 activation in more detail. Nevertheless, the properties and behavior of the inactive mutant of caspase-8, C360S-GFP, have suggested that it may prove to be a valuable tool for probing how caspase-8 interacts with the mitochondria after death receptor activation.
We had previously demonstrated that caspase-8 autoprocessing after Fas activation was reliant on an association of caspase-8 with the mitochondrial membrane, and that this association was dependent on the presence of mature cardiolipin.19 In the present study, we have identified a macromolecular complex of caspase-8 and BID on the mitochondrial membrane within which the cleavage of BID (tBID formation) occurs (Figure 8). Together, our results support the premise that Fas- or TRAIL-induced production of active caspase-8 on the mitochondria is necessary to target mitochondria-associated BID. Importantly, we have shown four key points to support this idea, namely, that caspase-8 was found in a complex with both its substrate (BID) and product (tBID) (Figures 5b and 6b), a larger proportion of mitochondria-bound BID was cleaved by caspase-8 compared with cytosolic BID (Figure 6d), incubation of isolated mitochondria with active caspase-8 was sufficient to induce MOMP (Figure 7a) and the amount of BID on the mitochondrial membrane was sufficient to support MOMP (Figure 7c). Overall, these results might explain why tBID is found exclusively on the mitochondrial membrane and why it, like p43-p10, has an affinity for cardiolipin. It should be noted that both full-length BID and tBID preferentially bound cardiolipin-containing liposomes in an in vitro assay (Supplementary Figure S7). In addition, we have also found that cells deficient in mature cardiolipin contain less mitochondria-bound BID (data not shown). These data suggested, in addition to the previously described model in which cardiolipin provides an activation platform for caspase-8, that cardiolipin is also necessary for the function and stabilization of the caspase-8/BID complex. A number of independent studies have observed the association of caspase-8 with the mitochondria, but the purpose of its localized production on the mitochondrial outer membrane has remained unknown.19, 28, 29, 30, 31, 32 Our results suggest that the formation of a death-inducing activation platform on the outer mitochondrial may be needed to effectively target sufficient amounts of BID for the induction of MOMP and to bypass the problem of weak caspase-8 activation at the DISC.
Schematic cartoon depicting a model for type II extrinsic apoptosis. The first step entails the initial activation of apoptosis by Fas or TRAIL, and the formation of the DISC. The next step is the stable insertion of active caspase-8 (p43-p10) into the mitochondrial membrane. In the third step, active caspase-8 cleaves BID within the native complex on the mitochondrial membrane, which results in the formation of tBID. Interestingly, after cleavage, tBID gains the ability to dissociate from the caspase-8/BID complex. Cleavage of BID exposes the BH3-domain, and may be the driving force behind tBID dissociation from the caspase-8/BID complex.25 tBID then shifts to separate complexes on the mitochondrial membrane where it interacts with BAK and/or BCL-XL to activate MOMP. Apoptogenic factors, such as cytochrome c and Smac/DIABLO, are released from the mitochondria, which leads to the activation of effector caspases and cell death
Recently, the presence of cardiolipin-enriched microdomains in the mitochondrial membrane had been described.33 In those experiments, Triton X-100 was used to solubilize the membranes, a detergent often used in the isolation of lipid rafts.34 Interestingly, we found that the caspase-8/BID complex was not detectable by 2D CNE/SDS-PAGE when Triton X-100 was used to solubilize the mitochondria (Supplementary Figure S8). Mitochondria-bound caspase-8 and BID were found solely in the Triton X-100-resistant membrane fraction (Supplementary Figure S8), a result that was consistent with the data presented in Figures 5 and 6, where higher amounts of active caspase-8 were found in DDM-resistant mitochondrial membranes. Collectively, these data would suggest that the caspase-8/BID complex localizes to mitochondrial membrane microdomains, and that disruption of these microdomains (i.e. by depletion of mature cardiolipin) inhibits complex formation and caspase-8 activation. However, the presence of mitochondrial lipid rafts is still a matter of debate and characterization of the caspase-8/BID complex and the role of caridolipin in its function requires further clarification.35 In summation, this novel caspase-8/BID mitochondrial complex concentrates the enzyme (caspase-8) and its substrate (BID) on the organelle where it is needed (mitochondria), thus providing a means by which the low levels of proteolytically active caspase-8 produced during type II apoptosis are able to efficiently engage the mitochondrial pathway.
Materials and Methods
Plasmids and expression vectors
The fluorescent-tagged caspase-8 expression constructs were generated by using human caspase-8 cDNA ligated into the KpnI and BamHI sites of pEGFP-N1 or pmCherry-N1 that expressed C-terminally tagged caspase-8. Caspase-8 point mutations were created by using a Site Directed Mutagenesis Kit (Stratagene). Primer sequences are available upon request. Mito-dsRed2 (Takara Bio Europe/Clontech, Saint-Germain-en-Laye, France) was a kind gift from Dr. K. Yee. The sequence targeted by siBID was GAAGACATCATCCGGAATATT. Non-targeting siRNA pool (Dharmacon; Thermo Fisher Scientific, Epsom, UK) was used as control. The hairpin sequence for shCaspase8 was CCGGGAATCACAGACTTTGGACAAACTCGAGTTTGTCCAAAGTCTGTGATTCTTTTT (Open Biosystems; Thermo Scientific, Epsom, UK). shCont HeLa and BCL-XL HeLa were described previously.19
Cell lines and reagents
The cervical carcinoma HeLa cell lines were cultured in DMEM supplemented with 10% FBS and 2 mM L-glutamine. Transfection of HeLa cells was performed using Lipofectamine 2000 (Invitrogen, Paisley, UK). BCL-XL, shCaspase-8 and shCont-stable HeLa cell lines were generated by transfection with pcDNA3/BCL-xL, pLKO.1/shCaspase-8 or pSUPER/shCont, respectively, and selected in G418 (0.5 mg/ml), except for shCaspase-8 (puromycin; 3 μg/ml). For siRNA transfection, unless otherwise stated, HeLa cells were transfected with 50 nM of the indicated siRNA, and the efficiency of the knockdown was validated by western blotting 48 h posttransfection. Recombinant human caspase-8 (R&D, Abingdon, UK) is the fully active form, which forms a stable tetramer composed of two dimers of the two subunits, p18 and p10. Antibodies used for immunoblot analyses were: anti-β-actin (Sigma, Dorset, UK), anti-α-tubulin (Sigma), anti-BCL-xL serum (a gift from Dr. Craig Thompson, University of Pennsylvania, Philadelphia, PA, USA), anti-BID (Cell Signaling, New England Biolabs (UK) Ltd., Hitchin, UK), anti-caspase-8 p18 domain (Alexis Biochemicals, Enzo Life Sciences (UK) Ltd., Exeter, UK), anti-cleaved caspase-8 (Cell Signaling), anti-cytochrome c (BD Pharmingen, Swindon, UK), anti-complex III (Reiske iron-sulfur protein) (Molecular Probes, Invitrogen, Paisley, UK), anti-complex I (ND6 protein) (Molecular Probes), anti-VDAC1 (Molecular Probes), anti-BAK (Upstate Biotechnology, Millipore, Jaffrey, NH, USA) and anti-GFP (Clontech).
Cell fractionation
HeLa cells were washed twice in cold PBS. Cells were then scraped into ice-cold PBS and spun at 600 × g for 10 min. Pellets were resuspended in mitochondrial isolation buffer (MIB) (200 mM Mannitol, 70 mM Sucrose, 1 mM EGTA, 10 mM HEPES; pH 7.4) supplemented with 1 mM dithiothreitol and 1X protease inhibitor cocktail (Sigma), and incubated on ice for 5 min. All subsequent steps were performed on ice. The cells were homogenized in a tight-fitting potter and an RZR 2051 overhead stirrer (Heidolph, Schwabach, Germany) at 1600 r.p.m. The nuclei and cell debris were separated and discarded by centrifugation at 600 × g for 10 min. The remaining supernatant was centrifuged at 10 000 × g for 10 min and the mitochondria-enriched fraction was washed once in MIB and then resuspended in a minimal volume of MIB. The mitochondrial and cytosolic protein concentrations were assessed using the Protein Assay Reagent (Bio-Rad, Hemel Hempstead, UK).
Confocal microscopy
In general, cells were grown in 6-well plates on glass coverslips. Cells were washed with PBS and fixed in 4% paraformaldehyde at room temperature. Coverslips were washed in PBS and mounted in immunofluorescence mounting medium (Vectashield; Vector Laboratories, Servion, Switzerland). Alternatively, cells were permeabilzed in Triton X-100, blocked with 10% FBS and incubated overnight with anti-cytochrome c (BD Pharmingen) in 5% BSA. Coverslips were incubated with Alexa Fluor 488 anti-mouse IgG (Molecular Probes) in 5% BSA, washed in PBS and mounted in immunofluorescence mounting medium containing DAPI. Specimens were analyzed at room temperature using a confocal microscope (Leica SP2 (DMIRBE) laser scanning confocal with Leica SP2 software) equipped with a 63x/1.32.ph3 oil HCX PL APO lens (Leica, Milton Keynes, UK).
Immunoprecipitation from isolated mitochondria
Isolated mitochondria were washed once by resuspending in 500 μl of MIB followed by centrifugation at 10 000 × g for 10 min. Pellets were resuspended in mitochondrial assay buffer (MAB) (120 mM KCl, 1 mM KH2PO4, 0.1 mM EGTA, 1 mM EDTA, 20 mM HEPES; pH 7.4). After resuspension, 1% CHAPS was added, and the mitochondrial membranes were incubated on ice for 5 min. Detergent-solubilized mitochondrial membranes were spun at 10 000 × g for 10 min. Supernatants were diluted to 0.5% CHAPS and incubated for 2 h at 4 °C with anti-GFP agarose beads (MBL International, Woburn, MA, USA). Beads were washed three times in MAB+0.5% CHAPS and resuspended in SDS sample buffer before western blot analysis.
Caspase-8 activity measurements
Isolated mitochondria were washed once in MIB followed by centrifugation at 10 000 × g for 10 min. Pellets were resuspended in MIB and incubated for 1 h at room temperature in the dark with Caspase-Glo 8 Reagent (Promega, Southampton, UK). The relative light unit (RLU) produced by caspase-8 cleavage of benzyloxycarbonyl-Leu-Glu-Thr-Asp (zLETD)-aminoluciferin was recorded using GLOMAX (Promega) software on a Veritas Microplate Luminometer (Promega).
Cell death assays and caspase-8 processing
At 24 h posttransfection, cells were harvested and washed in PBS. Pellets were resuspended in PBS containing 2 μg/ml propidium iodide (PI) (Molecular Probes, Invitrogen). The number of GFP-positive/PI positive cells was quantified by flow cytometry using a BD FACSCalibur system with CellQuest software (Becton Dickinson, Swindon, UK). Alternatively, pellets were resuspended directly in SDS sample buffer for analysis by SDS-PAGE.
In vitro cytochrome c release assays
Isolated mitochondria were resuspended in reaction buffer (125 mM KCl, 1 mM KH2PO4, 2.5 mM MgCl2, 0.2 mM EGTA, 5 mM HEPES, 0.1% BSA; pH 7.4) supplemented with 1X protease inhibitor cocktail (Sigma). Mitochondria were incubated for 5 min at room temperature with 10 mM succinate and 10 μM rotenone. Recombinant protein was added at the stated concentration and incubated for 15 min at 37 °C. Mitochondria were collected by centrifugation at 10 000 × g for 10 min. Supernatants and pellets were solubilized in SDS sample buffer and analyzed by western blotting. Alternatively, pellets were washed and resuspended in reaction buffer and incubated for 1 h at room temperature in the dark with Caspase-Glo 8 Reagent (Promega) to measure LETDase activity.
2D CNE/SDS-PAGE
Isolated mitochondria were washed once by resuspending in 500 μl of MIB followed by centrifugation at 10 000 × g for 10 min. Pellets were resuspended in mitochondrial resuspension buffer (50 mM NaCl, 50 mM imidazole/HCl, 1 mM EDTA; pH 7.1). In all, 2.5 g/g dodecyl-β-D-maltoside (Invitrogen) or 3.0 g/g Triton X-100 (Sigma) was added, and solubilization was allowed to proceed for 5–10 min on ice. Samples were spun at 100 000 × g for 17 min at 4 °C using an Optima MAX-XP ultracentrifuge (Beckman Coulter, High Wycombe, UK). The supernatant was removed and 4X sample buffer (Invitrogen) added. Pellets were solubilized directly in SDS-PAGE sample buffer. The solubilized proteins were loaded onto a 4–16% Native-PAGE precast gel (Invitrogen) and run at 4 °C. Where appropriate, fluorescence was measured using the Odyssey Infared Imaging System (Li-Cor Biosciences, Cambridge, UK) and Odyssey V1.2 software (Li-Cor). The gel lanes were excised and denatured in LDS sample buffer supplemented with 4% β-mercaptoethanol before being loaded onto a 4–12% Bis-Tris ZOOM gel (Invitrogen). Proteins were then electroblotted from the gel onto PVDF membranes for immunoblotting.
Liposome preparation and analysis
To analyze the binding of full-length BID and tBID to cardiolipin, these proteins were incubated with artificial liposomes either containing or lacking cardiolipin. The lipid composition of liposomes was based on the composition of mitochondrial contact sites as previously reported.36 Liposomes containing cardiolipin consisted of 41 mol% PC, 22 mol% PE, 8 mol% PI, 9 mol% cholesterol and 20 mol% cardiolipin. In the liposomes lacking cardiolipin, we increased both the PC and PE content to compensate for the lack of cardiolipin, resulting in a composition of 53 mol% PC, 30 mol% PE, 8 mol% PI and 9 mol% cholesterol. Bovine liver PC, PE and PI were from Avanti Polar Lipids (Alabaster, AL, USA); bovine heart cardiolipin and cholesterol were purchased from Sigma. For the preparation of the liposomes, lipids were dissolved in chloroform, dried under a stream of nitrogen and resuspended in binding buffer (100 mM NaCl, 2 mM MgCl2, 10 mM Tris; pH 7.1). The lipid mixture was passed 11 times through a 400 nm polycarbonate membrane of a Mini-Extruder (Avanti Polar Lipids) to form large unilamellar vesicles. Following precipitation and wash of liposomes at 100 000 × g for 15 min at 4 °C, the presence or absence of cardiolipin was confirmed by MS analysis. The indicated proteins were incubated with the liposomes in binding buffer at 37 °C for 30 min, and liposomes were then centrifuged, washed and lysed for protein analysis.
Abbreviations
- TRAIL:
-
tumor necrosis factor-related apoptosis-inducing ligand
- TNF:
-
tumor necrosis factor
- MOMP:
-
mitochondrial outer membrane permeabilization
- DISC:
-
death-inducing signalling complex
- DED:
-
death effector domain
- zVAD-fmk:
-
benzyloxycarbonyl-Val-Ala-DL-Asp-fluoromethylketone
- DDM:
-
dodecyl-β-D-maltoside
- CNE:
-
clear native electrophoresis
- VDAC1:
-
voltage dependent anion channel 1
- TOM20:
-
translocase of outer membrane 20
- cyt. c:
-
cytochrome c
- rC8:
-
recombinant active caspase-8
- CL:
-
cardiolipin
References
van Loo G, Saelens X, van Gurp M, MacFarlane M, Martin SJ, Vandenabeele P . The role of mitochondrial factors in apoptosis: a Russian roulette with more than one bullet. Cell Death Differ 2002; 9: 1031–1042.
Wang X . The expanding role of mitochondria in apoptosis. Genes Dev 2001; 15: 2922–2933.
Leber B, Lin J, Andrews DW . Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis 2007; 12: 897–911.
Chipuk JE, Green DR . How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? 2008; 18: 157–164.
Peter ME, Budd RC, Desbarats J, Hedrick SM, Hueber AO, Newell MK et al. The CD95 receptor: apoptosis revisited. Cell 2007; 129: 447–450.
Boldin MP, Goncharov TM, Goltsev YV, Wallach D . Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell 1996; 85: 803–815.
Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM . An induced proximity model for caspase-8 activation. J Biol Chem 1998; 273: 2926–2930.
Martin DA, Siegel RM, Zheng L, Lenardo MJ . Membrane oligomerization and cleavage activates the caspase-8 (FLICE/MACHalpha1) death signal. J Biol Chem 1998; 273: 4345–4349.
Shi Y . Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell 2002; 9: 459–470.
Chen M, Orozco A, Spencer DM, Wang J . Activation of initiator caspases through a stable dimeric intermediate. J Biol Chem 2002; 277: 50761–50767.
Keller N, Mares J, Zerbe O, Grutter MG . Structural and biochemical studies on procaspase-8: new insights on initiator caspase activation. Structure 2009; 17: 438–448.
Hughes MA, Harper N, Butterworth M, Cain K, Cohen GM, MacFarlane M . Reconstitution of the death-inducing signaling complex reveals a substrate switch that determines CD95-mediated death or survival. Mol Cell 2009; 35: 265–279.
Chang DW, Xing Z, Capacio VL, Peter ME, Yang X . Interdimer processing mechanism of procaspase-8 activation. EMBO J 2003; 22: 4132–4142.
Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM et al. A unified model for apical caspase activation. Mol Cell 2003; 11: 529–541.
Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli KJ et al. Two CD95 (APO-1/Fas) signaling pathways. EMBO J 1998; 17: 1675–1687.
Lee KH, Feig C, Tchikov V, Schickel R, Hallas C, Schutze S et al. The role of receptor internalization in CD95 signaling. EMBO J 2006; 25: 1009–1023.
Luo X, Budihardjo I, Zou H, Slaughter C, Wang X . Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 1998; 94: 481–490.
Li H, Zhu H, Xu CJ, Yuan J . Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 1998; 94: 491–501.
Gonzalvez F, Schug ZT, Houtkooper RH, MacKenzie ED, Brooks DG, Wanders RJ et al. Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J Cell Biol 2008; 183: 681–696.
Schug ZT, Gottlieb E . Cardiolipin acts as a mitochondrial signalling platform to launch apoptosis. Biochim Biophys Acta 2009; 1788: 2022–2031.
Zou H, Li Y, Liu X, Wang X . An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem 1999; 274: 11549–11556.
Tinel A, Tschopp J . The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 2004; 304: 843–846.
Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 2009; 137: 721–735.
Wang C, Youle RJ . The Role of Mitochondria in Apoptosis. Annu Rev Genet 2009; 43: 95–118.
Gross A, Yin X-M, Wang K, Wei MC, Jockel J, Milliman C et al. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1/Fas death. J Biol Chem 1999; 274: 1156–1163.
Yin X-M, Wang K, Gross A, Zhao Y, Zinkel S, Klocke B et al. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature 1999; 400: 886–891.
Scaffidi C, Krammer PH, Peter ME . Isolation and analysis of components of CD95 (APO-1/Fas) death-inducing signaling complex. Methods 1999; 17: 287–291.
Stegh AH, Barnhart BC, Volkland J, Algeciras-Schimnich A, Ke N, Reed JC et al. Inactivation of Caspase-8 on mitochondria of Bcl-xL-expressing MCF7-Fas cells. Role for the bifunctional apoptosis regulator protein. J Biol Chem 2002; 277: 4351–4360.
Stegh AH, Herrmann H, Lampel S, Weisenberger D, Andra K, Seper M et al. Identification of the cytolinker plectin as a major early in vivo substrate for caspase 8 during CD95- and tumor necrosis factor receptor-mediated apoptosis. Mol Cell Biol 2000; 20: 5665–5679.
Chandra D, Choy G, Deng X, Bhatia B, Daniel P, Tang DG . Association of active caspase 8 with the mitochondrial membrane during apoptosis: potential roles in cleaving BAP31 and caspase 3 and mediating mitochondrion-endoplasmic reticulum cross talk in etoposide-induced cell death. Mol Cell Biol 2004; 24: 6592–6607.
Qin ZH, Wang Y, Kikly KK, Sapp E, Kegel KB, Aronin N et al. Pro-caspase-8 is predominantly localized in mitochondria and released into cytoplasm upon apoptotic stimulation. J Biol Chem 2001; 276: 8079–8086.
Milovic-Holm K, Krieghoff E, Jensen K, Will H, Hofmann TG . FLASH links the CD95 signaling pathway to the cell nucleus and nuclear bodies. EMBO J 2007; 26: 391–401.
Sorice M, Manganelli V, Matarrese P, Tinari A, Misasi R, Malorni W et al. Cardiolipin-enriched raft-like microdomains are essential activating platforms for apoptotic signals on mitochondria. FEBS Lett 2009; 583: 2447–2450.
Waugh MG, Hsuan JJ . Preparation of membrane rafts. Methods Mol Biol 2009; 462: 403–414.
Zheng YZ, Berg KB, Foster LJ . Mitochondria do not contain lipid rafts, and lipid rafts do not contain mitochondrial proteins. J Lipid Res 2009; 50: 988–998.
Lutter M, Fang M, Luo X, Nishijima M, Xie X, Wang X . Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat Cell Biol 2000; 2: 754–761.
Acknowledgements
This work was supported by Cancer Research UK. F.M. Vaz was supported by grants from the Prinses Beatrix Fonds (no WAR05-0126) and the Barth syndrome foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by SJ Martin
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Schug, Z., Gonzalvez, F., Houtkooper, R. et al. BID is cleaved by caspase-8 within a native complex on the mitochondrial membrane. Cell Death Differ 18, 538–548 (2011). https://doi.org/10.1038/cdd.2010.135
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2010.135
Keywords
This article is cited by
-
The E3 ubiquitin ligase Itch regulates death receptor and cholesterol trafficking to affect TRAIL-mediated apoptosis
Cell Death & Disease (2024)
-
Combined inhibition of histone deacetylase and cytidine deaminase improves epigenetic potency of decitabine in colorectal adenocarcinomas
Clinical Epigenetics (2023)
-
Harnessing TRAIL-induced cell death for cancer therapy: a long walk with thrilling discoveries
Cell Death & Differentiation (2023)
-
Checkpoint inhibitor immunotherapy diminishes oocyte number and quality in mice
Nature Cancer (2022)
-
Arylamine N-acetyltransferase 1 deficiency inhibits drug-induced cell death in breast cancer cells: switch from cytochrome C-dependent apoptosis to necroptosis
Breast Cancer Research and Treatment (2022)