Abstract
Background:
To estimate the accuracy of one-step nucleic acid amplification (OSNA) assay as an intra-operative sentinel lymph node biopsy (SLNB) for sentinel lymph node (SLN) metastasis in breast cancer.
Methods:
PubMed, Cochrane Library and Web of Science databases were searched by two independent reviewers to retrieve literature with per-patient analysis. The deadline was up until December 2016. A meta-analysis was performed using STATA, Meta-Disc, and Revman software. A random-effects model was used and subgroup analysis was carried out to identify possible sources of heterogeneity.
Results:
According to the inclusion criteria, 2833 patients from 12 studies were included in this meta-analysis. The pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) and the area under the sROC curve (AUC) for detecting SLN metastasis were 0.87 (95% CI 0.81–0.91), 0.92 (95% CI 0.86–0.95), 10.65 (95% CI 6.18–18.34), 0.14 (95% CI 0.10–0.20), 75.08 (95% CI 37.77–149.22) and 0.94 (95% CI 0.91–0.95), respectively.
Conclusions:
The present study adds the evidence that OSNA assay is an accurate molecular diagnostic tool for intra-operatively detecting SLN metastasis in breast cancer. One-step nucleic acid amplification assay might be introduced into clinical usage for replacing traditional intro-operative diagnostic methods of SLNB.
Similar content being viewed by others
Main
One-step nucleic acid amplification (OSNA) assay is a molecular detection technique that might be introduced into clinical usage for intra-operative detection of sentinel lymph node (SLN) metastasis in breast cancer. Many studies focusing on OSNA assay have been published; however, so far, there is no clear consensus on the diagnosis performances of OSNA assay. The present study evaluates the performance of OSNA assay for intra-operative diagnosis of SLN metastasis in breast cancer on a per-patient basis. With high sensitivity, specificity, diagnostic odds ratio (DOR) and AUC, we figured out that OSNA assay is an accurate diagnostic tool with an advantage over traditional intro-operative methods of sentinel lymph node biopsy (SLNB).
Breast cancer is the most common malignancy in women. One-third of all breast cancer patients will suffer from metastasis, which is the main cause of mortality (Molloy et al, 2012). Axillary lymph node dissection (ALND) is a traditional clinical procedure to identify axillary lymph node metastasis in breast cancer; however, since ALND is an invasive method, patients who have undergone ALND might suffer from unpleasant complications (Wang et al, 2011). In order to provide evidence for the surgery plan without patients’ invasive pains, SLNB is widely recommended for intra-operative diagnosis of patients with early-stage breast cancers. By intra-operative evaluation of SLNB, patients with positive nodes are spared a second surgical procedure as well as patients with clinically node-negative avoid undesirable ALND (Kumar et al, 2012).
Essentially, SLN analysis includes intra-operative evaluation and post-operative pathological examination. Multistep formalin-fixed tissue sections stained by haematoxylin and eosin (H&E) with or without immunohistochemistry (IHC) is usually accepted in post-operative pathological detection (Tamaki et al, 2012), yet this method, based on formalin-fixed tissue sections, is not appropriate to intra-operative assessment. The frozen section (FS) and imprint cytology (IC) are the most commonly used techniques for intra-operative SLN evaluation. Unfortunately, the two methods suffered from low sensitivity ranging from 57 to 74% compared with post-operative pathological examination (Khaddage et al, 2011), in addition, they are hampered by variable accuracy and nonstandard operational approach.
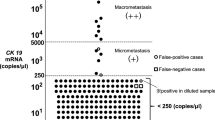
With the advantage of standardisation, reliability and repetitiveness, molecular diagnostic techniques for intra-operative detection of SLN won the favour of many pathologists and clinicians. One-step nucleic acid amplification (Sysmex, Kobe, Japan) assay is established as an automatic technology for accurately identifying lymph node metastasis by quantitatively evaluating the number of cytokeratin 19 (CK19) mRNA copies (Osako et al, 2011). According to the cut-off levels defined previously by Tsujimoto et al (2007), the CK19 mRNA copy numbers larger than 5000 per μl of dilution homogenate are classified as (++), copy numbers between 250 and 5000 per μl as (+), and copy numbers less than 250 per μl as (−). The (++) and (+) results of OSNA assay, demonstrated by clinical trials, are well consistent with SLN macrometastasis and micrometastasis, while (−) judgement of OSNA assay represents that the lymph node has no tumour deposits or only isolated tumour cells.
So far there is no clear consensus on the diagnosis performance of OSNA for detecting SLN metastasis in patients with breast cancer. A meta-analysis performed by Tiernan et al suggested that the sensitivity and specificity of OSNA is 0.87 and 0.98 (Tiernan et al, 2014), whereas the sensitivity of 0.85 and specificity of 0.92 were observed by a systematic review conducted by Huxley et al (2015), included studies of them were published before December 2012 and were mainly based on a per-node analysis. Considering that numerous studies whose data analysis is obtained on a per-patient basis have been published and that the intra-operative detection is performed on each patient, evaluating the performance of OSNA assay on a per-patient basis is of great clinical significance (Chen et al, 2013); however, there is no meta-analysis with sufficient data to address this problem. This present meta-analysis was conducted aiming to investigate the intra-operative diagnosis performance of OSNA for detecting SLN metastasis based on a per-patient analysis, which will definitely complement previous researches.
Materials and methods
Search strategy and study selection
To retrieve all relevant papers published up to 31 December 2016, three databases including PubMed, Cochrane Library and Web of Science were searched by two independent reviewers (SHI F and ZHANG Q). Search terms were defined as follows: ‘one-step nucleic acid amplification’ or OSNA, and breast cancer or breast neoplasms or breast carcinoma or breast malignant tumour or mammary cancer or mammary neoplasm or mammary carcinoma or mammary malignant tumour. The detailed search strategies were shown in Supplementary Table 1. The references of all related papers were also checked for potential studies.
All articles were screened according to the inclusion and exclusion criteria by two independent reviewers (SHI F and ZHANG Q). The inclusion criteria were as follows: (1) patients included in the studies were diagnosed with breast cancer; (2) patients did not undergo any new adjuvant therapy or radiotherapy; (3) the diagnostic purpose was to detect SLN metastasis; (4) the specimens detected were fresh SLNs; (5) the reference standard method for the detection of SLN metastasis was post-operative histopathology; (6) the same patient was detected by OSNA and post-operative histopathology together; (7) detailed method of post-operative histopathology was described in detail; (8) data analysis of study was based on per-patient; and (9) extracted data were available for calculating true-positive, false-positive, false-negative, and true-negative values. The exclusion criteria of this study were set as follows: (1) Non-English literatures; (2) Non-clinical research literatures including basic experiments, reviews, conference abstracts and letters to journal editors; (3) The analysis of study was based on a per-node model; and (4) The data published were not sufficient to form 2 × 2 tables.
Data extraction and quality assessment
For each eligible study, the following information was extracted by two independent reviewers (SHI F and LIANG Z): first author, year of publication, type of study design, country, number of patients, mean age of patients, number of slices used for pathological examination, thickness of slices detected by pathological examination, interval of serial slices detected by pathological examination, reference standard method, whether IHC for CK19 was brought into the reference standard method, the turnaround time of OSNA assay for detecting one node, and whether the result was adjusted for excluding tissue allocation bias (TAB). Parameters such as true-positive, false-positive, false-negative, and true-negative values were placed into 2 × 2 tables. Conflicting results were evaluated by a third-party researcher (LIU X) and resolved consensually.
To assess the quality of included literature, Quality Assessment of diagnostic Accuracy Studies 2 (QUADAS-2) (Whiting et al, 2011) was conducted by two independent reviewers (SHI F and ZHANG M). QUADAS-2 was an evidence-based quality assessment tool that consisted of four domains: patient selection, index test, reference standard, and flow and timing.
Statistical analysis
First, threshold effect and heterogeneity were measured. If the P-value of Spearman correlation coefficient was more than 0.05, there was no threshold effect, whether a heterogeneity caused by non-threshold effect existed was further analysed. The heterogeneity was evaluated by P-value of Cochran-Q test and I-square statistic. If P>0.05 and I2⩽50%, studies were thought to be free from the significant heterogeneity, and a fixed effects model was performed to calculate the pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR) and DOR. If P<0.05 or I2>50%, which meant the existence of statistical heterogeneity, a random-effect model was constructed subsequently and a subgroup analysis was further performed to identify possible sources of heterogeneity. The eight covariates for subgroup analysis were as follows: (1) race of patients, (2) the number of patients, (3) the number of slices used for pathological examination, (4) the thicknesses of slices detected by pathological examination, (5) the interval of slices detected by pathological examination, (6) detailed approaches of reference pathology, (7) utilising IHC for CK19 in reference method, and (8) adjusting the result for TAB. If P-value was more than 0.05 and I2 was not greater than 50% in one subgroup, the according covariate was considered as source of heterogeneity. The pooled sensitivity, specificity, PLR, NLR and DOR, along with their 95% confidence intervals (CIs) were computed and graphically displayed using forest plots. An sROC curve was constructed to access the superior diagnostic accuracy for OSNA, and AUC was calculated; the higher the AUC was, the higher the diagnostic value was. Publication bias was assessed by Deeks’ funnel plot asymmetry test. The meta-analysis was undertaken using STATA13.0 (StataCorp, College Station, TX, USA) and Meta-Disc 1.4 (Unit of Clinical Biostatistics, Ramo e Cajal Hospital, Madrid, Spain). Quality assessments of included studies were carried out with RevMan 5.3 (Revman, the Cochrane Collaboration). P<0.05 was considered of statistical significance.
Results
Study selection
According to the search strategy, a total of 394 articles from PubMed, Cochrane Library and Web of Science databases were retrieved. No new articles were found after screening the references. Among these articles, 113 were excluded because of duplication and 269 articles were excluded for they did not meet the inclusion criteria: 117 articles were not related to the diagnosis effort of OSNA for breast cancer; 7 articles were with adjuvant therapy or radiotherapy; the subjects of 8 articles were non-SLNs rather than SLNs; 1 paper was non-English literature; 95 papers were non-clinical research literature; 36 articles were excluded because data analysis was not based on a per-patient analysis; 5 articles were excluded because data were not sufficient to form 2 × 2 tables. Finally, 12 articles were included in this meta-analysis (Figure 1).
Study characteristics
The 12 eligible articles (Choi et al, 2010; Khaddage et al, 2011; Snook et al, 2011; Le Frere-Belda et al, 2012; Vegue et al, 2012; Wang et al, 2012; Chen et al, 2013; Heilmann et al, 2013; Jimbo et al, 2013; Bettington et al, 2014; Pathmanathan et al, 2014; and Mokhtar et al, 2016) containing a total of 2833 patients with a mean sample size of 218 (range: 35–772) were included in this meta-analysis. All studies were designed by prospective methods. There were two adopted approaches of reference pathology: the detection of formalin-fixed tissue sections with H&E staining (8/12), as well as combined detection of H&E and IHC (4/12). Among the four studies taken with IHC, three studies utilised IHC for CK19 while one utilised IHC for anti-cytokeratin antibody AE1 and AE3. One whole node was commonly cut into four parts and divided equally between OSNA assay and pathology. The thicknesses of the slices used for pathological examination were mostly more than 1 mm, and the interval between the slices for pathological examination in most studies was 200 μm. Three studies adjusted the result for excluding TAB. The turnaround time of OSNA assay for detecting one node was all less than 40 min (Table 1).
Risk of bias and quality assessment
Deek’s funnel chart was used to analyse publication bias (Supplementary Figure 1). The funnel chart was symmetrical and P>0.05, suggesting no significant publication bias. The results of the QUADAS-2 (Supplementary Figure 2) suggested that the risk of bias was low and the overall quality of included studies were of moderate to high.
Diagnostic effect
The pooled sensitivity and specificity (Figure 2) of OSNA assay for detecting SLN metastasis were 0.87(95% CI 0.81–0.91) and 0.92 (95% CI 0.86–0.95), respectively. The pooled PLR and NLR (Figure 3) were 10.65 (95% CI 6.18–18.34) and 0.14 (95% CI 0.10–0.20), respectively. The pooled DOR (Supplementary Figure 3) was 75.08 (95% CI 37.77–149.22) and AUC (Figure 4) was 0.94 (95% CI 0.91–0.95), respectively.
Heterogeneity and subgroup analysis
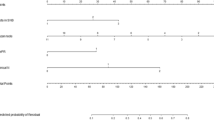
In the threshold analysis, spearman correlation coefficient was −0.074 and P-value was 0.820, indicating no threshold effect. But a significant heterogeneity caused by non-threshold effect was observed. As shown in Figure 3, I2 of sensitivity was 59.26%, and I2 of specificity was 92.74%. A random-effects model was used since the existence of high heterogeneity. The subgroup analysis was further conducted to identify possible sources of heterogeneity. The result of subgroup analysis showed that the number and interval of slices used for pathology might be responsible for sensitivity heterogeneity, and the major source of specificity heterogeneity was not found (Table 2).
Discussion
Numerous studies have focused on the diagnosis performance of OSNA assay for intra-operatively detecting SLN metastasis in breast cancer. However, no consistent conclusion has ever been made up to present. A meta-analysis based on a per-node analysis suggested the sensitivity and specificity of OSNA is 0.87 and 0.98 (Tiernan et al, 2014), whereas the sensitivity of 0.85 and specificity of 0.92 were observed by Huxley N et al (Huxley et al, 2015); however, as Huxley N et al only included five studies based on per-patient analysis, the small number of included studies limited the quality of evidence. Moreover, papers included in previous systematic reviews were all published before December 2012. The aim of this study is to investigate the diagnosis effort of OSNA assay on a per-patient basis and all related papers before 2017 were included in our work. The pooled sensitivity, specificity, PLR, NLR, DOR and AUC of OSNA for detecting SLN metastases are 0.87(95% CI 0.81–0.91), 0.92 (95% CI 0.86–0.95), 10.65 (95% CI 6.48–18.34), 0.14 (95% CI 0.10–0.20), 75.08 (95% CI 37.77–149.22) and 0.94 (95% CI 0.91–0.95). This meta-analysis demonstrates that OSNA assay is a reliable and accurate diagnosis method for SLNs.
Discordant cases between OSNA and reference pathology are analysed in detail. The false-positive results may be potentially due to TAB. TAB refers to a sample bias yielding because of the uneven distribution of metastatic foci throughout the node (Chen et al, 2013). In all studies included in our meta-analysis, one whole node is divided into several alternate slices and a same slice cannot be detected by both OSNA assay and pathological examination; thus a TAB inevitably yields. Conventional pathological method potentially underestimates the degree of tumour burden as only a few proportion of node sample is detected (Castellano et al, 2012). Considering that OSNA assay detects a greater portion or even a whole of node in clinical practice, the missed diagnosis of tumour burden is well avoided (Choi et al, 2010; Snook et al, 2011). Moreover, subgroup analysis of our study (sensitivity: 0.91 vs 0.83, specificity: 0.94 vs 0.90 and DOR: 160.21 vs 53.37) shows that the presence of TAB affects the result of research, suggesting more studies with rigorous design are needed in this area. The false-negative results usually exist owing to low CK19 expression. CK19 is an epithelial marker highly expressed in up to approximately 98% of breast cancers, which means approximately 2% of breast cancer patients lack CK19 expression; moreover, certain subtypes such as metaplastic cancers, lobular cancers, luminal (ER+) cancers and triple-negative phenotype breast cancer are more prone to the lack of CK19 expression (Abd El-Rehim et al, 2004; Bettington et al, 2014). As CK19 is the single molecular marker used in OSNA assay, low CK19 expression of nodes may result in a false-negative result. In our work, a better DOR of OSNA assay (160.21 vs 53.37) was observed when studies adopted IHC for CK19; so IHC for CK19 is recommended to be carried out to examine the CK19 expression in patients with the above subtypes and OSNA should not be omitted for patients lacking CK19 expression. Meanwhile, a greater sensitivity by using multiple markers together rather than a single marker has been observed by some findings (Snook et al, 2011; Le Frere-Belda et al, 2012); so the accuracy of intro-operative diagnosis of SLNs may be improved by using multiple markers such as mammaglobin and CK19.
The SLNB, a widely accepted clinical staging method for cancers, provides evidence for the appropriate selection of surgical treatment and reduces pain and surgical cost for each patient with breast cancer (Tamaki et al, 2012). Frozen section and TIC are the most commonly used intro-operative detection method of SLNB; however, the value of them is still controversial. These techniques are hampered by a rather low sensitivity and a no unified standardisation. A meta-analysis assessing intra-operative FS for SLNs showed that the pooled sensitivity of FS was 73% (Liu et al, 2011). The poor sensitivity is owing to a limited amount of tissue detected by FS, as well as samples suffering from destructive, freezing, and compression artefacts (Olson et al, 2007). Tew et al reported that the pooled sensitivity of IC was 63% (Tew et al, 2005). IC technique fails to provide information about size of a metastatic deposit that can assist intra-operative decision-making (Bettington et al, 2014). Our results show that the sensitivity of OSNA assay is 86%, indicating that OSNA assay is superior to FS and IC for intro-operative diagnosis of SLN metastasis.
With a shorter turnaround time, automated operating procedure and quantitative results, OSNA assay has obvious advantages in comparison with post-operative pathology with formalin-fixed tissue sections. Four SLNs can be detected by OSNA assay at the same time (Wang et al, 2012), whereas only a single SLN can be evaluated by pathology at once. The turnaround time of OSNA assay was all less than 40 min for detecting one node and ranges from 50 to 62 min for detecting four nodes, demonstrating that OSNA assay can meet the time demand for rapid diagnosis of SLNs and can be incorporated into intra-operative clinical setting. Moreover, the turnaround time can be reduced with an accumulating experience of technicians. Besides less turnaround time, OSNA assay is easier to operate compared with labour-intensive pathology. As RNA purification is not required and mixing of reagents or samples for CK19 mRNA amplification is performed by an automatic device, the manual processes in OSNA assay are quite easy (Khaddage et al, 2011); hence manipulation errors made by technicians are greatly reduced. The results of pathological examination depend on the subjective judgments of pathologist, which means accuracy of results may be influenced by experience and expertise of pathologists (Chen et al, 2013). As OSNA assay quantitatively evaluates the number of CK19 mRNA copies and outcomes correlate well with the number of cancer cells in a node as well as all procedure of OSNA follows a uniform protocol, the result is objective and repeatable.
Although OSNA assay has many advantages, it still has some inherent limitations. One-step nucleic acid amplification assay does not provide histological information, such as extra capsular extension and occult pathology, which is associated with the patient’s prognosis (Bettington et al, 2014). Besides, this method would be hampered by the lack of morphological correlation information such as the location of metastasis, size, pattern of spread and extra nodal spread, which is important information for predicting non-SLN involvement and can be used in guiding the selection of optimal therapeutic regimen (Bernet et al, 2011; Pathmanathan et al, 2014).
Considering the huge benefits derived from OSNA assay, OSNA is still proposed to introduce into intra-operative clinical setting. One-step nucleic acid amplification assay has been recommended by NICE guidance for diagnosis of SLN metastasis in breast cancer (Chaudhry et al, 2014). By introducing highly sensitive OSNA assay into intra-operative clinical usage, a recall for the second surgical procedure is avoided; thus the number of hospitalisations and related costs is reduced.
Our study includes 12 articles with OSNA based on a per-patient analysis and makes up for deficiencies in previous studies. However, there are two limitations in our work. Firstly, a statistical heterogeneity of specificity was observed in our study, however, the major source of specificity heterogeneity was not found. Secondly, non-English studies were excluded from this meta-analysis which might cause selection bias.
In conclusion, our meta-analysis adds the evidence that OSNA assay is a rapid and accurate molecular method for intra-operatively detecting SLN metastasis in breast cancer. One-step nucleic acid amplification assay can be introduced into routine clinical setting and replace FS as well as IC in intra-operative SLNB.
Change history
10 October 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Abd El-Rehim DM, Pinder SE, Paish CE, Bell J, Blamey RW, Robertson JF, Nicholson RI, Ellis IO (2004) Expression of luminal and basal cytokeratins in human breast carcinoma. J pathol 203 (2): 661–671.
Bernet L, Cano R, Martinez M, Duenas B, Matias-Guiu X, Morell L, Palacios J, Rezola R, Robles-Frias M, Ruiz I, Velasco A, Vieites B, Sevilla F, Torro J, Medrano J, Ballester B (2011) Diagnosis of the sentinel lymph node in breast cancer: a reproducible molecular method: a multicentric Spanish study. Histopathology 58 (6): 863–869.
Bettington M, Lakhani SR, Ung OA (2014) Is the one-step nucleic acid amplification assay better for intra-operative assessment of breast sentinel nodes? ANZ J Surg 84 (10): 725–729.
Castellano I, Macri L, Deambrogio C, Balmativola D, Bussone R, Ala A, Coluccia C, Sapino A (2012) Reliability of whole sentinel lymph node analysis by one-step nucleic acid amplification for intraoperative diagnosis of breast cancer metastases. Ann surg 255 (2): 334–342.
Chaudhry A, Williams S, Cook J, Jenkins M, Sohail M, Calder C, Winters ZE, Rayter Z (2014) The real-time intra-operative evaluation of sentinel lymph nodes in breast cancer patients using One Step Nucleic Acid Amplification (OSNA) and implications for clinical decision-making. Eur j surg oncol 40 (2): 150–157.
Chen JJ, Chen JY, Yang BL, Yang WT, Shao ZM, Wang YS, Ou YT, Cao XC, Liu YH, Wu J (2013) Comparison of molecular analysis and touch imprint cytology for the intraoperative evaluation of sentinel lymph nodes in primary breast cancer: results of the China Breast Cancer Clinical Study Group (CBCSG) 001c trial. Eur j surg oncol 39 (5): 442–449.
Choi YL, Ahn SK, Bae YK, Park IA, Min JW, Lee KW, Jin HB, Han W, Choi JE, Cho EY (2010) One-step Nucleic Acid Amplification (OSNA): intraoperative rapid molecular diagnostic method for the detection of sentinel lymph node metastases in breast cancer patients in Korean cohort. J Breast Cancer 13 (4): 366.
Heilmann T, Mathiak M, Hofmann J, Mundhenke C, van Mackelenbergh M, Alkatout I, Wenners A, Eckmann-Scholz C, Schem C (2013) Intra-operative use of one-step nucleic acid amplification (OSNA) for detection of the tumor load of sentinel lymph nodes in breast cancer patients. J cancer res clin oncol 139 (10): 1649–1655.
Huxley N, Jones-Hughes T, Coelho H, Snowsill T, Cooper C, Meng Y, Hyde C, Mujica-Mota R (2015) A systematic review and economic evaluation of intraoperative tests [RD-100i one-step nucleic acid amplification (OSNA) system and Metasin test] for detecting sentinel lymph node metastases in breast cancer. Health technol assess (Winchester, England) 19 (2): v–xxv, 1–215.
Jimbo K, Kinoshita T, Suzuki J, Asaga S, Hojo T, Yoshida M, Tsuda H (2013) Sentinel and nonsentinel lymph node assessment using a combination of one-step nucleic acid amplification and conventional histological examination. Breast 22 (6): 1194–1199.
Khaddage A, Berremila SA, Forest F, Clemenson A, Bouteille C, Seffert P, Peoc'h M (2011) Implementation of molecular intra-operative assessment of sentinel lymph node in breast cancer. Anticancer res 31 (2): 585–590.
Kumar A, Puri R, Gadgil PV, Jatoi I (2012) Sentinel lymph node biopsy in primary breast cancer: window to management of the axilla. World j surg 36 (7): 1453–1459.
Le Frere-Belda MA, Bats AS, Gillaizeau F, Poulet B, Clough KB, Nos C, Peoc'h M, Seffert P, Bouteille C, Leroux A, Guillemin F, Blanc-Fournier C, Crouet H, Arnould L, Cuisenier J, Penault-Llorca F, Gimbergues P, Jacquemier J, Houvenaeghel G, Chatellier G, Lecuru F (2012) Diagnostic performance of one-step nucleic acid amplification for intraoperative sentinel node metastasis detection in breast cancer patients. Int J Cancer 130 (10): 2377–2386.
Liu LC, Lang JE, Lu Y, Roe D, Hwang SE, Ewing CA, Esserman LJ, Morita E, Treseler P, Leong SP (2011) Intraoperative frozen section analysis of sentinel lymph nodes in breast cancer patients: a meta-analysis and single-institution experience. Cancer 117 (2): 250–258.
Mokhtar M, Tadokoro Y, Nakagawa M, Morimoto M, Takechi H, Kondo K, Tangoku A (2016) Triple assessment of sentinel lymph node metastasis in early breast cancer using preoperative CTLG, intraoperative fluorescence navigation and OSNA. Breast Cancer 23 (2): 202–210.
Molloy TJ, Roepman P, Naume B, van't Veer LJ (2012) A prognostic gene expression profile that predicts circulating tumor cell presence in breast cancer patients. PloS one 7 (2): e32426.
Olson TP, Harter J, Munoz A, Mahvi DM, Breslin T (2007) Frozen section analysis for intraoperative margin assessment during breast-conserving surgery results in low rates of re-excision and local recurrence. Ann surg oncol 14 (10): 2953–2960.
Osako T, Iwase T, Kimura K, Yamashita K, Horii R, Akiyama F (2011) Accurate staging of axillary lymph nodes from breast cancer patients using a novel molecular method. Br J Cancer 105 (8): 1197–1202.
Pathmanathan N, Renthawa J, French JR, Edstrom-Elder E, Hall G, Mahajan H, Teh C, Bilous MA (2014) Intraoperative sentinel lymph node assessment in breast cancer: a comparison of rapid diagnostic method based on CK19 mRNA expression and imprint cytology. ANZ J Surg 84 (10): 730–734.
Snook KL, Layer GT, Jackson PA, de Vries CS, Shousha S, Sinnett HD, Nigar E, Singhal H, Chia Y, Cunnick G, Kissin MW (2011) Multicentre evaluation of intraoperative molecular analysis of sentinel lymph nodes in breast carcinoma. Br j surg 98 (4): 527–535.
Tamaki Y, Sato N, Homma K, Takabatake D, Nishimura R, Tsujimoto M, Yoshidome K, Tsuda H, Kinoshita T, Kato H, Taniyama K, Kamio T, Nakamura S, Akiyama F, Noguchi S (2012) Routine clinical use of the one-step nucleic acid amplification assay for detection of sentinel lymph node metastases in breast cancer patients: results of a multicenter study in Japan. Cancer 118 (14): 3477–3483.
Tew K, Irwig L, Matthews A, Crowe P, Macaskill P (2005) Meta-analysis of sentinel node imprint cytology in breast cancer. Br j surg 92 (9): 1068–1080.
Tiernan JP, Verghese ET, Nair A, Pathak S, Kim B, White J, Thygesen H, Horgan K, Hanby AM (2014) Systematic review and meta-analysis of cytokeratin 19-based one-step nucleic acid amplification versus histopathology for sentinel lymph node assessment in breast cancer. Br j surg 101 (4): 298–306.
Tsujimoto M, Nakabayashi K, Yoshidome K, Kaneko T, Iwase T, Akiyama F, Kato Y, Tsuda H, Ueda S, Sato K, Tamaki Y, Noguchi S, Kataoka TR, Nakajima H, Komoike Y, Inaji H, Tsugawa K, Suzuki K, Nakamura S, Daitoh M, Otomo Y, Matsuura N (2007) One-step nucleic acid amplification for intraoperative detection of lymph node metastasis in breast cancer patients. Clin cancer res 13 (16): 4807–4816.
Vegue LB, Rojo F, Hardisson D, Iturriagagoitia AC, Panades MJ, Velasco A, Bonet EL, Munoz RC, Polo L (2012) Comparison of molecular analysis and histopathology for axillary lymph node staging in primary breast cancer: results of the B-CLOSER-I study. Diagn mol pathol 21 (2): 69–76.
Wang YS, Ou-yang T, Wu J, Liu YH, Cao XC, Sun X, Fu L, Liao N, Yang WT, Zhong WX, Lu AP (2012) Comparative study of one-step nucleic acid amplification assay, frozen section, and touch imprint cytology for intraoperative assessment of breast sentinel lymph node in Chinese patients. Cancer Sci 103 (11): 1989–1993.
Wang Z, Wu LC, Chen JQ (2011) Sentinel lymph node biopsy compared with axillary lymph node dissection in early breast cancer: a meta-analysis. Breast cancer research treat 129 (3): 675–689.
Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM (2011) QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann intern med 155 (8): 529–536.
Acknowledgements
This study was supported by Technology Project of the Department of Health of Jilin Province (Grant No. 20165044).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Shi, F., Zhang, Q., Liang, Z. et al. One-step nucleic acid amplification assay is an accurate technique for sentinel lymph node biopsy of breast cancer patients: a meta-analysis. Br J Cancer 117, 1185–1191 (2017). https://doi.org/10.1038/bjc.2017.262
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.262
Keywords
This article is cited by
-
Accuracy of One-Step Nucleic Acid Amplification in Detecting Lymph Node Metastases in Endometrial Cancer
Pathology & Oncology Research (2020)
-
Sentinel lymph node biopsy in breast cancer—an updated overview
European Surgery (2020)
-
External validation of a prognostic model based on total tumor load of sentinel lymph node for early breast cancer patients
Breast Cancer Research and Treatment (2020)
-
The new TNM-based staging of breast cancer
Virchows Archiv (2018)